Improve reporting practices for quantitative neuroscience data
06 August 2020
A team of HBP researchers from the University of Oslo has created a database of quantitative information about cellular parameters in the murine basal ganglia.
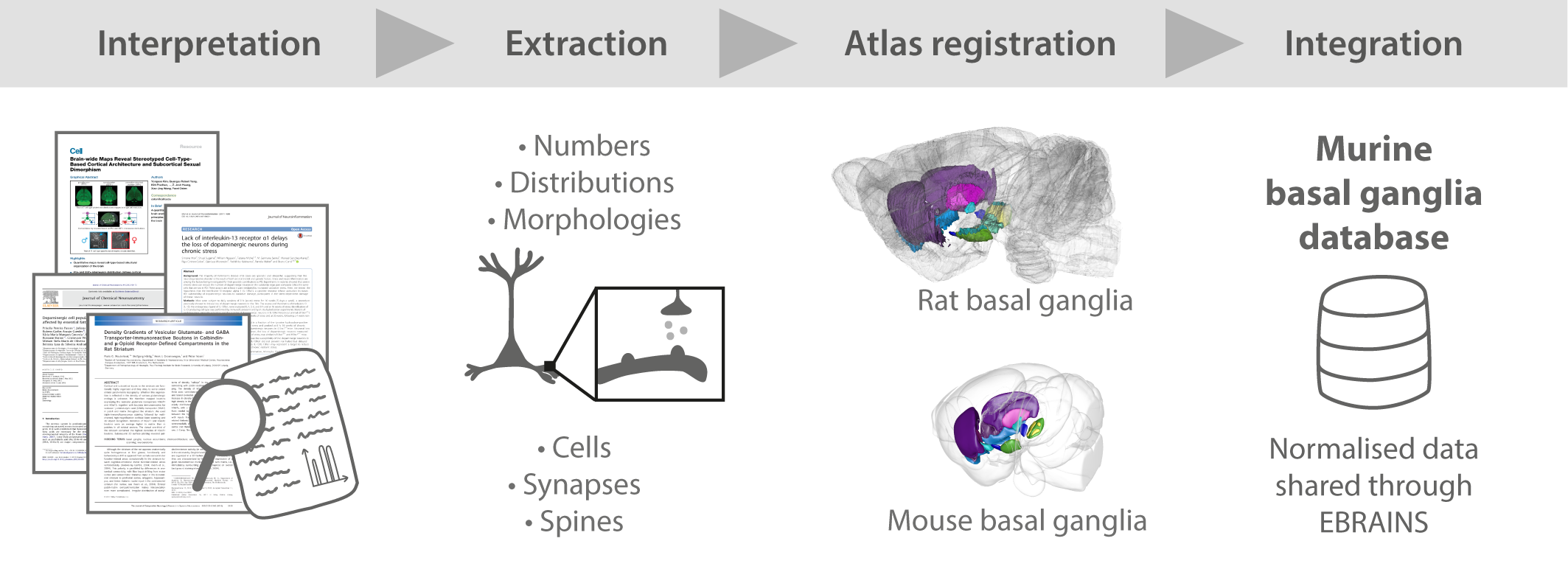
The researchers’ work, published in Nature: Scientific Data, provides the most thorough collection of quantitative neuroanatomical data from the basal ganglia to date. The data set has been made available as a downloadable resource from the EBRAINS Knowledge Graph.
A vibrant user community is developing around EBRAINS and is helping to transform the neuroscience field into a data-driven science. Scientists submitting their data to EBRAINS receive data management support, long term storage and citable DOIs. Curated neuroscience datasets can be searched through the EBRAINS Knowledge Graph and visually explored through the EBRAINS brain atlases.
EBRAINS users can share their data through the FAIR data service (making data Findable, Accessible, Interoperable, and Re-usable) and thereby obtain greater exposure of their research, or access shared data assets and boost their productivity.
For the neurosciences, the new EBRAINS data services have been added to Nature Scientific Data’s list of recommended repositories.
In a recent blog post for the Springer Nature Research Data portal, HBP PhD student and first author Ingvild Bjerke explains the need to improve reporting practices for quantitative neuroscience data: “Improved reporting practices are urgently needed to ensure that published information can have a value beyond the narrative of a specific publication.”
The researchers hope that their shared database will make it easier for neuroscientists to find, extract, and compare quantitative data.
Further reading:
Improve reporting practices for quantitative neuroscience data
Database of literature derived cellular measurements from the murine basal ganglia



