Brain Atlases
Contact us
Contact Brain Atlases Support here or email us at support@ebrains.eu
Community
Join the Brain Atlas Space in the EBRAINS Community
Brain atlases provide spatial reference systems for neuroscience that allow navigation, characterisation and analysis of information based on anatomical location. A 3D interactive viewer to explore brain atlases, as well as tools for integration into workflows, spatial anchoring, and atlas-based data analysis have been developed by the Human Brain Project (HBP) and are accessible via EBRAINS.
An atlas of the brain defines the shape and location of brain regions in a common coordinate space. The brain atlases developed in the HBP are openly available through the EBRAINS research infrastructure, and include detailed atlases of the human brain and the rat brain, as well as a new atlas of the macaque brain.
Explore the human, mouse, rat and (pre-released) monkey atlases in 3D using the interactive viewer siibra-explorer
interactive viewer siibra-explorer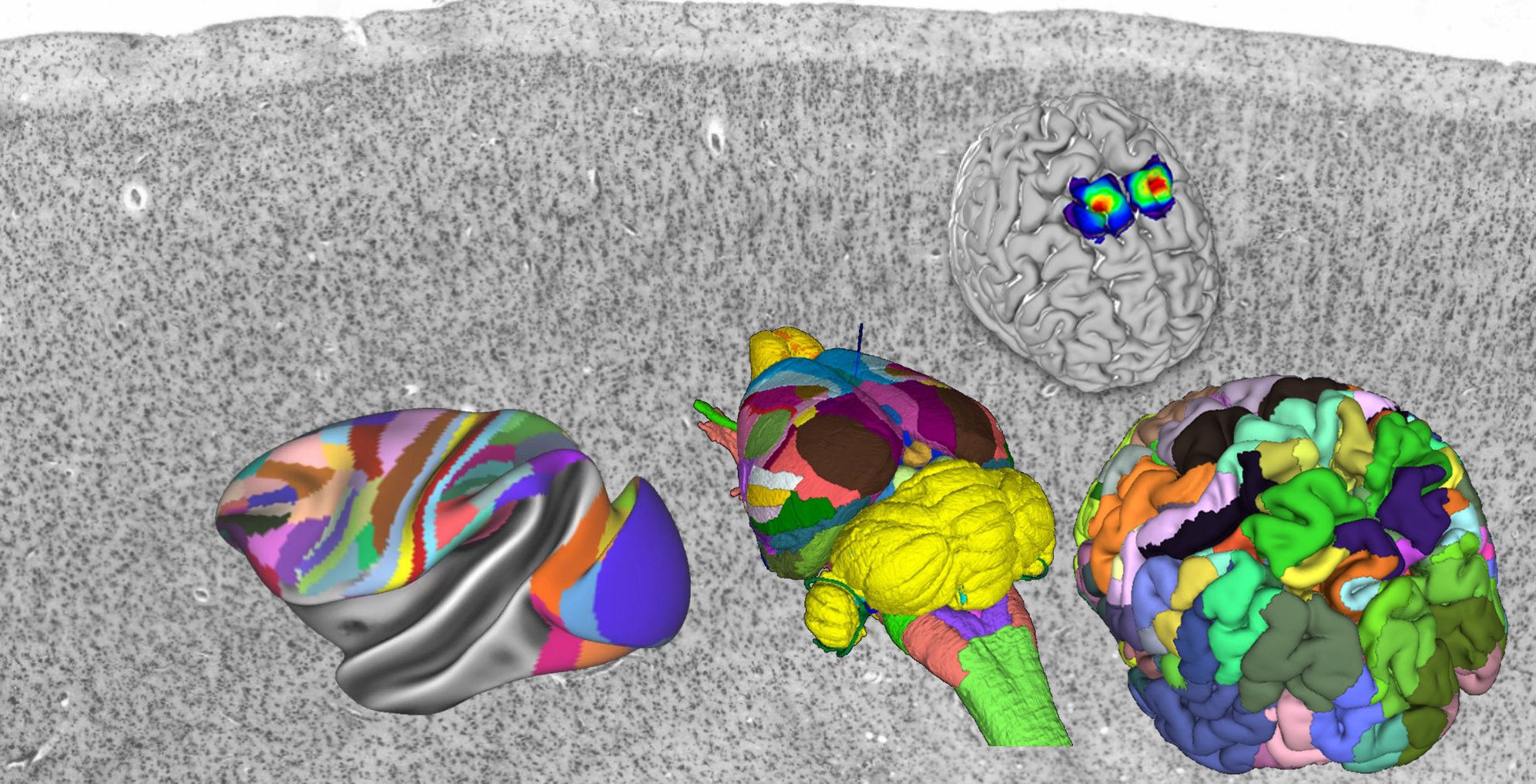
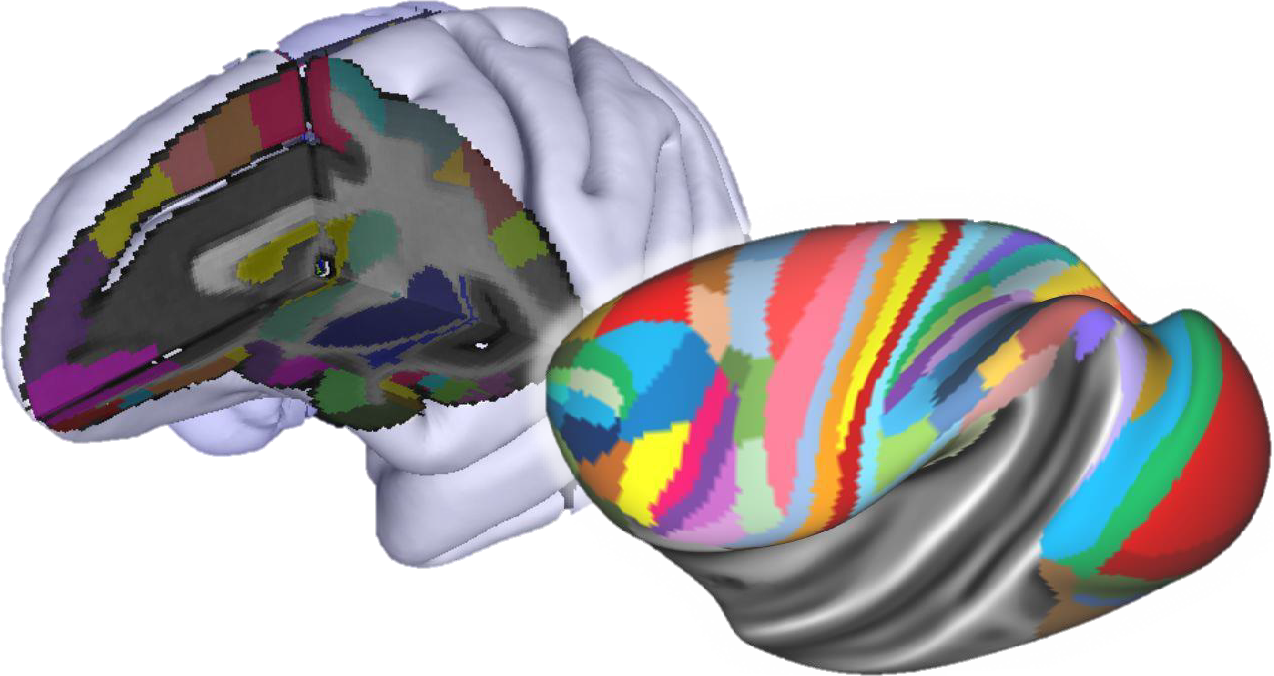
The multilevel Human Brain Atlas captures information about brain variability in different subjects and accumulates knowledge from various neuroscientific sources both at the microscopic and macroscopic scales, including cytoarchitecture, structural and functional connectivity, fibre architecture, and chemoarchitecture. Based on microstructural principles, it builds links between MRI-based reference spaces to the microscopic BigBrain model. The easiest way to explore the multilevel Human Brain Atlas is by using the interactive atlas viewer, siibra-explorer.
Find out more about the Multilevel Human Brain Atlas here.
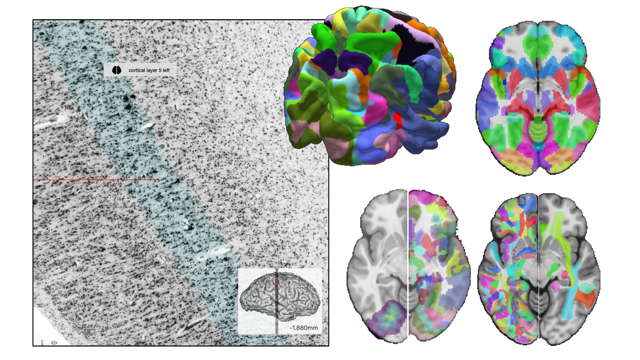
The multilevel Non-Human Primate (NHP) brain atlas will integrate aspects of brain organisation revealed by complementary structural, functional and connectivity-based datasets defined across multiple spatial and temporal resolutions. It is based on a reference template that represents an average of high-resolution structural (T1, T2) magnetic resonance images and CT scans of 10 macaque monkey brains. The NHP brain atlas will provide maps of distinct cyto- and receptor architectonically identified areas in the macaque monkey brain, and thus links with the brain’s organisation at the cellular and molecular levels. The easiest way to explore the Non-Human Primate Brain Atlas is by using the interactive atlas viewer, siibra-explorer.
The Waxholm Space (WHS) Rat Brain Atlas contains comprehensive anatomical delineations of the brain regions and fibre tracts of an adult Sprague Dawley rat brain. The atlas was defined and generated by experts using high-resolution ex-vivo magnetic resonance and diffusion tensor imaging. The easiest way to explore the Waxholm Space Rat Brain Atlas is by using the interactive atlas viewer, siibra-explorer.

Find out more about the Waxholm Space Rat Brain Atlas here.
The Allen Mouse Brain Common Coordinate Framework (CCF) is used as a basis for combining and comparing data from mouse brains and is an essential tool for understanding the structure and function of the mouse brain at the molecular, cellular and behavioural levels. It is defined in a population-averaged image volume using serial two-photon tomography. The easiest way to explore the Allen Mouse Brain Atlas is by using the interactive atlas viewer, siibra-explorer.
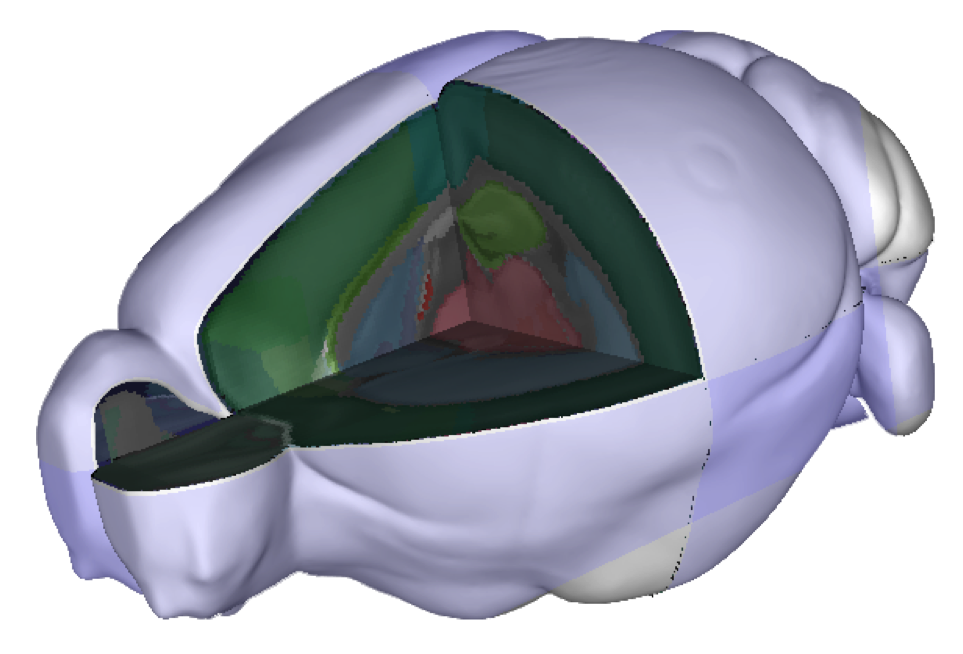
Find out more about the Allen Mouse Brain Common Coordinate Space here.
Atlases Tools and Services
The atlas tools and services are available through the EBRAINS infrastructure.
EBRAINS atlases can be interactively explored in full detail using the 3D atlas viewer siibra-explorer right from a web browser. The viewer supports browsing 3D reference brain templates at MRI and full microscopic resolutions, accessing multimodal data features from the EBRAINS Knowledge Graph by their location in the brain, and exploring links between spatial scales and modalities. The viewer also provides basic annotation capabilities and plugins for interactive data analysis.
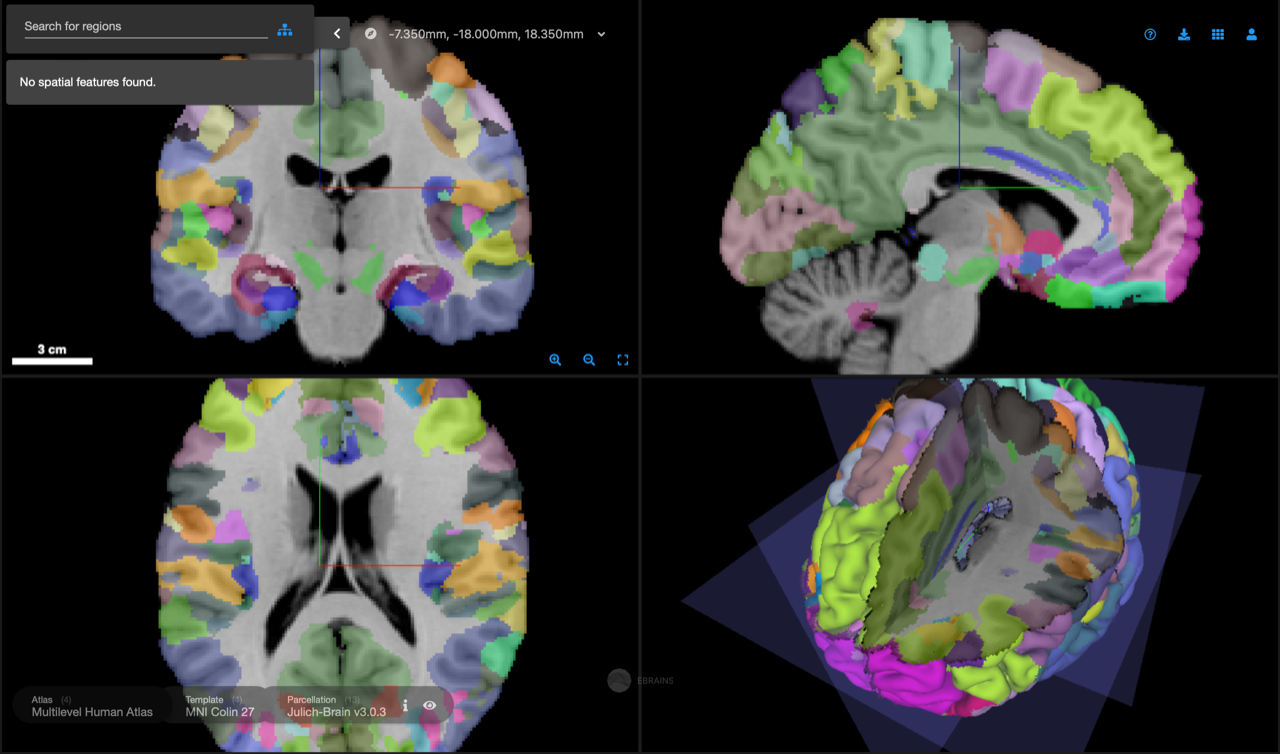
Access siibra-explorer
siibra-python is a comprehensive Python client for integrating EBRAINS brain atlases into scripts, reproducible workflows and applications. The library allows to access all multiscale parcellation maps and reference templates included in EBRAINS, querying multimodal data features, and performing atlas-related analyses such as anatomical assignment of coordinates or images. The software is maintained on GitHub, with regular releases on pypi, a growing documentation and a set of interactive tutorials.
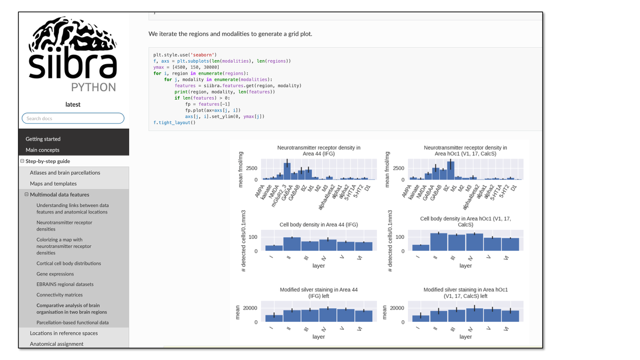
Access siibra-python on GitHub
QuickNII and VisuAlign allow rapid and efficient mouse and rat 2D brain image registration to a 3D volumetric reference atlas.
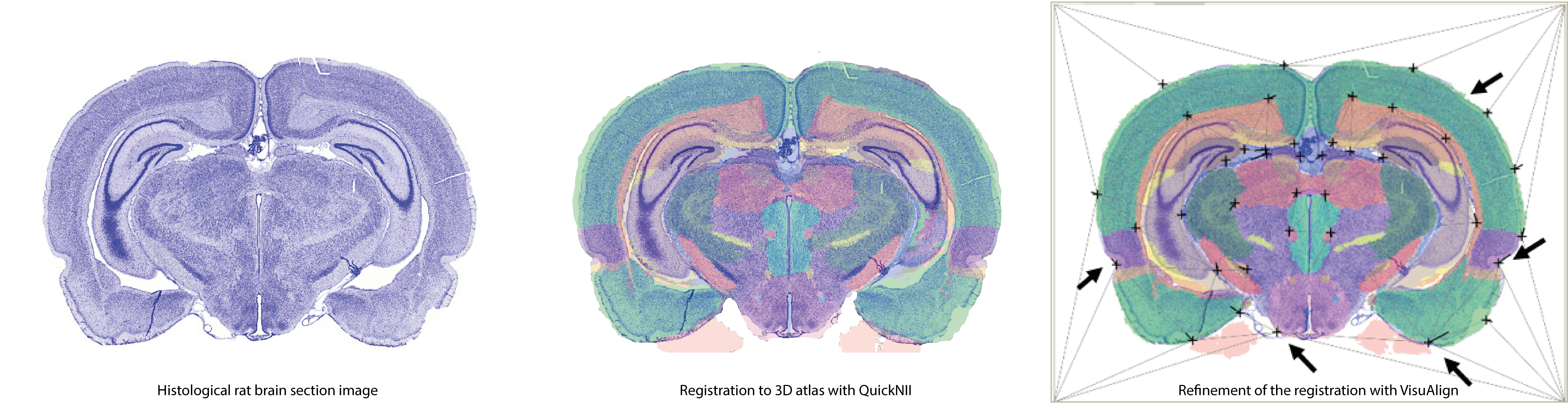
Download QuickNII and VisuAlign from Nitric.
Read more about QuickNII and VisuAlign
voluba (volumetric brain anchoring) is a browser-based tool for the interactive linear alignment of volumes of interest (VOIs) from brain imaging experiments to a high-resolution 3D reference brain model.
Read more about voluba here.
Launch voluba
The QUINT analytical workflow enables an atlas-based analysis of extracted features from histological image sections from the rodent brain by using 3D reference atlases.
Read more about the QUINT workflow here.
Get user support and download QUINT
JuGEx (Julich-Brain Gene Expression) is a tool for gene expression analysis in human brain atlas regions.
Read more about JuGEx here.
Open the JuGEx plugin in siibra-explorer