QUINT: New workflow for registration and analysis of experimental 2D image data to brain atlases
16 January 2020
How can we better understand the interplay of thousands of cells in brains of animal models, and in both healthy and diseased brains? Researchers worldwide are generating images showing the positions of specific populations of cells in brain sections as well as some of the key substances in these cells. Some of these substances reveal the nature of the cells (neurons, astrocytes, microglia, oligodendrocytes, ..), other reveal the function of cells (excitatory, inhibitory), and yet others give us information about which systems the cells participate in (dopamine, acetylcholine, ..). Images from models of disease mechanisms may also reveal information about pathological hallmarks, such as beta-amyloid plaques in Alzheimer’s disease.
By gathering all this information in a common reference map, we are in a better position to decipher the complex interactions that occur between these features, ultimately increasing our understanding of brain function and disease. The QUINT workflow enables just this integration in a common reference system. HBP teams from the University of Oslo and EMBL in Heidelberg have developed new software tools, and together with experts on Alzheimer’s disease from the University of Leipzig, University of Munich, and the Fraunhofer Institute in Halle, present the QUINT workflow for 2D brain image analysis, published in Frontiers in Neuroinformatics.
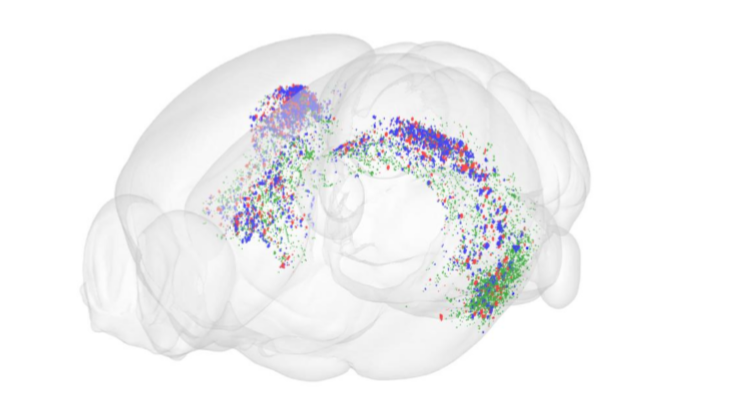 Image: Sharon Yates. Location of human amyloid in blue, mouse amyloid in red and pyroglutamate modified amyloid in green, in Hippocampal regions of transgenic Tg2573 mouse brain.
Image: Sharon Yates. Location of human amyloid in blue, mouse amyloid in red and pyroglutamate modified amyloid in green, in Hippocampal regions of transgenic Tg2573 mouse brain.
"We are driven by the needs of the neuroscience community”, says Dr. Maja Puchades, University of Oslo. “Many different tools exists but this is the first time a whole workflow can be applied to large image data collections from different laboratories, and it doesn’t require any coding skills”. The tools and workflow developed are entirely open access and are already being tested by several laboratories outside of the HBP, including the Korea Brain Research Institute (Daegu) and the Kaczorowski Lab at The Jackson Laboratory (Bar Harbor). One of the tools is included in the tools catalogue of NIH's Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative - Cell Census Network (BICCN). All tools will be accessible through the EBRAINS platform.
Publications and software availability:
Scientific contact:
Dr. Maja A. Puchades
Nesys laboratory, Institute of Basic Medical Sciences
University of Oslo, Norway
Email: m.a.puchades@medisin.uio.no
Dr. Anna Kreshuk
Cell biology and Biophysics
EMBL Heidelberg, Germany
Email: anna.kreshuk@embl.de
Support:
Access The HBP Research Infrastructure (RI) and Receive Support



