Simulations
Contact us
Contact Simulations Support here or email us at support@ebrains.eu
Community
Join the Simulation Engines and Data-Driven Simulations Community Spaces in the EBRAINS Community
Publications
Find related publications here.
What is Brain Simulation?
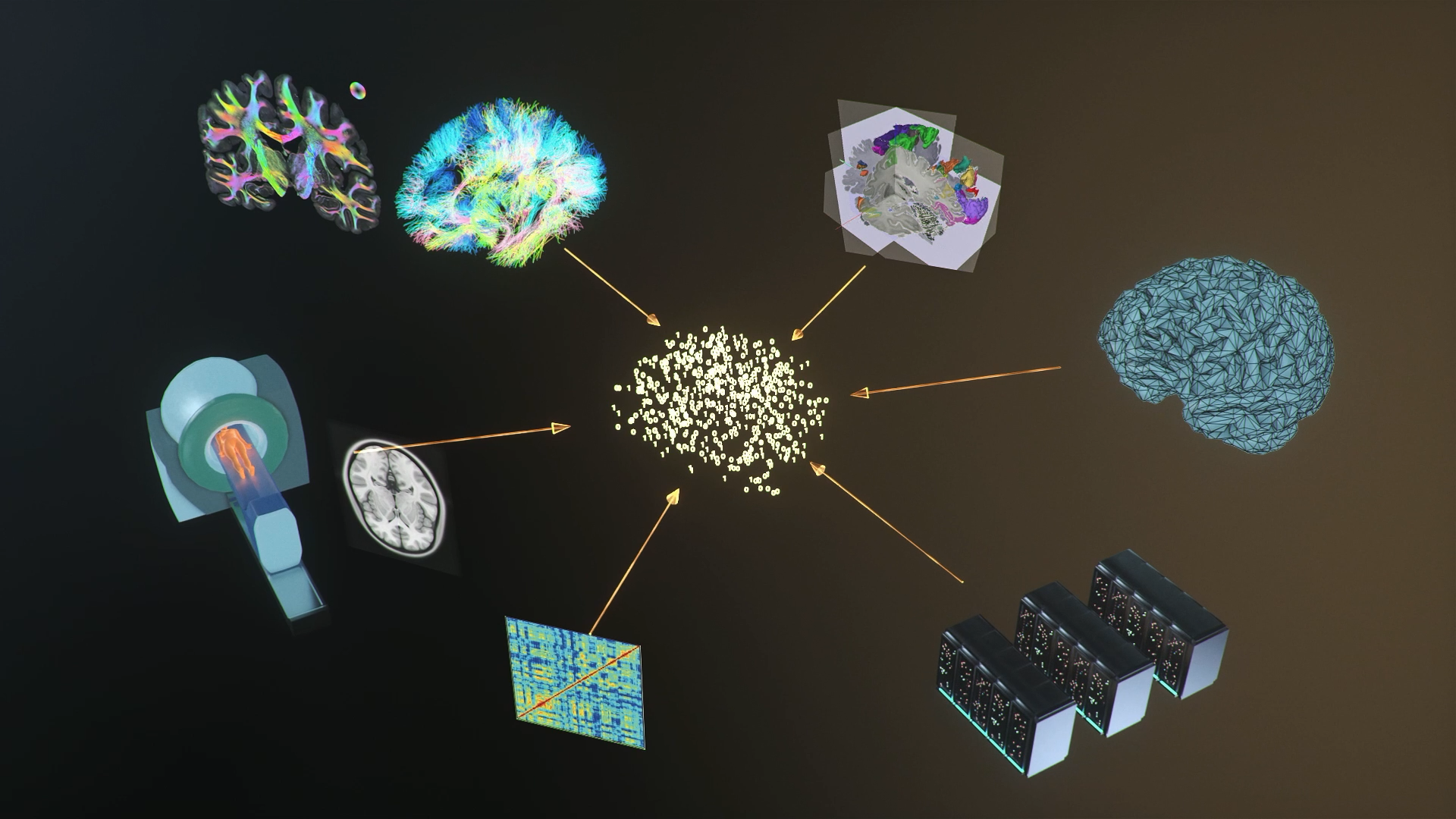
Brain simulation tools and services which are developed within the HBP offer technical solutions for brain researchers to conduct sustainable simulation studies and build upon prior work, and the means to share their results. The tools and services provide integrated workflows for model creation, simulation and validation, including data analysis and visualisation. The simulation engines cover the entire spectrum of levels of description ranging from molecular and subcellular to cellular, network and whole brain level. Multi-simulator interaction enables coupling of simulation engines operating at different levels of description and thus allows for concurrent simulation of detailed models of smaller scope embedded into more abstract models of larger scope.
Visit the EBRAINS website on simulations to learn more about the simulation engines integrated in EBRAINS.
Simulation tools, services and workflows
Whole-brain level simulation
Network level simulation
Cellular level simulation
Molecular and subcellular level simulation
Multi-scale co-simulation
Data analysis and visualisation
How simulation can support your research
Whole-brain level simulation
To simulate whole brains, the white-matter axon fibre bundle network, the so-called structural connectome, is reconstructed to define the model network. To reduce complexity and computational demands, the activity of brain regions is typically not simulated by networks of individual neurons. Instead, mean-field theory is used to replicate the main dynamics of large groups of neurons. The resulting models of the different networks are connected by the weights and time delays of reconstructed structural connectomes. Whole-brain simulations have been used to explain several empirically observed phenomena, such as the emergence in fMRI of functional networks, the spread of epileptic seizures or relationships between phenomena observed in fMRI and electrical neural activity over several temporal scales. In the HBP/EBRAINS, the simulation tools at the whole brain level were developed around the framework of The Virtual Brain (TVB). Two Facility Hubs support The Virtual Brain software and The Virtual Brain Cloud end-to-end workflow services.
The Virtual Brain (TVB) is an open-source platform for constructing and simulating personalised brain network models. The TVB-on-EBRAINS ecosystem includes a variety of prepackaged modules, integrated simulation tools, pipelines and data sets for easy and immediate use on EBRAINS. Process your large cohort databases and use these results to develop potential medical treatments, therapies or diagnostic procedures.
Network level simulation
Brain function emerges at the network level. Even basic functions such as the initial processing of visual input require the interaction of many neurons in different brain areas. Network-level simulation technology enables investigation of such brain-scale networks, covering multiple brain areas at the resolution of single neurons and synapses. The direct link to brain function makes network-level simulation attractive for AI and robotics applications. To enable brain-scale simulations, the technology needs to be extremely scalable and capable of exploiting modern supercomputers. The simplifications that underpin network-level simulation technology have also inspired the design of neuromorphic systems.
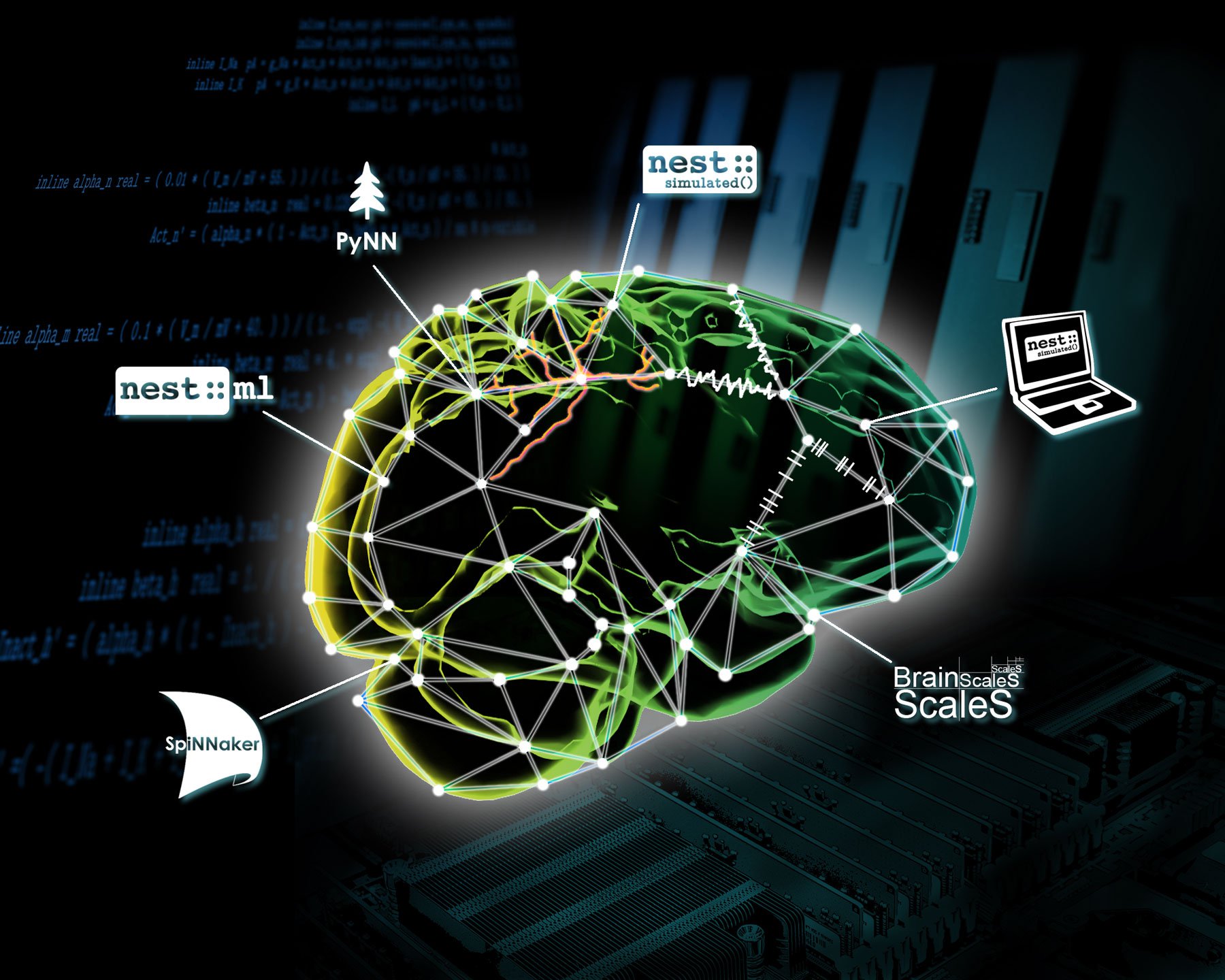
NEST is a simulator for spiking neural network models that focuses on the dynamics, size and structure of neural systems, rather than on the exact morphology of individual neurons. It is ideal for networks of any size, including models of information processing (e.g. in the visual or auditory cortex of mammals), models of network activity dynamics (e.g. laminar cortical networks or balanced random networks) and models of learning and plasticity. NEST is openly available for download. Visit https://www.nest-simulator.org/ for more details.
NEST Desktop is a web-based application which provides a graphical user interface for NEST Simulator. NEST Desktop enables the rapid construction, parametrisation and instrumentation of neuronal network models. It offers a graphical user interface for construction of networks, running simulations in NEST and applying visualisation to support the analysis of simulation results. The main goal of NEST Desktop is to offer an install-free (web-based), interactive visual tool for classroom use in computational neuroscience courses as well as young scientist in computational neuroscience. Visit https://nest-desktop.readthedocs.io/en/latest/ for more details.
NESTML is a user-friendly domain-specific language, that enables neuroscientists to write custom neuron and synapse models quickly and easily. This is achieved without compromising on performance, by automatic source-code generation of optimized simulation code for a given target platform, such as NEST Simulator or SpiNNaker. The language itself as well as the associated toolchain are lightweight, modular and extensible. Algorithms can be implemented using the built-in procedural language, and differential equations can be written as a simple string in mathematical notation. Verifying correctness of the model and generating target platform code are fully automated by the associated toolchain, written in Python. For differential equations, symbolic mathematics routines compute exact solutions where possible, or select an appropriate numeric solver otherwise. NESTML comes packaged with over two dozen neuron and synaptic plasticity models. Visit https://nestml.readthedocs.io/en/latest/ for more details.
PyNN is both (i) a Python API for simulator-independent modelling and simulation of spiking neuronal networks and (ii) a reference implementation of this API for the NEST, NEURON and Brian simulators, distributed as a Python package (library). Implementations of the API are also available for the SpiNNaker and BrainScaleS neuromorphic computing systems. The principal advantage of PyNN in EBRAINS is that the same code can be used both on software simulators such as NEST and on neuromorphic hardware such as SpiNNaker. PyNN can be installed on HPC systems and in Jupyter notebooks, and is pre-installed on the Neuromorphic Computing platform and the Neurorobotics platform.
Network models described using the PyNN API can be simulated on the BrainScaleS and SpiNNaker platforms, either interactively via Jupyter notebooks, or in batch mode by submitting jobs to a central job queue. Three different clients - a web app, a Python client, and a command-line application - are available to anyone with an EBRAINS account. These tools can all be used both to submit jobs and to view/download results. The app is currently available in the EBRAINS Collaboratory and the Python client, which includes the command-line application, can be downloaded from the Python Package Index.