The scientific case for brain simulations
03 June 2019
HBP scientists argue for brain simulators as “mathematical observatories” for neuroscience
Simulations of large-scale networks of neurons are a key element in the European Human Brain Project (HBP). In a new perspective article scientists from the HBP argue why such simulations are indispensable for bridging the scales between the neuron and system levels in the brain. The authors describe the need for open general-purpose simulation engines that can run a multitude of different candidate models of the brain at different levels of biological detail. Comparing predictions derived from such simulations with experimental data will allow systematic testing and refinement of models in a loop between computational and experimental neuroscience.
The article has been published as a featured perspective in the leading journal Neuron and can be read here.
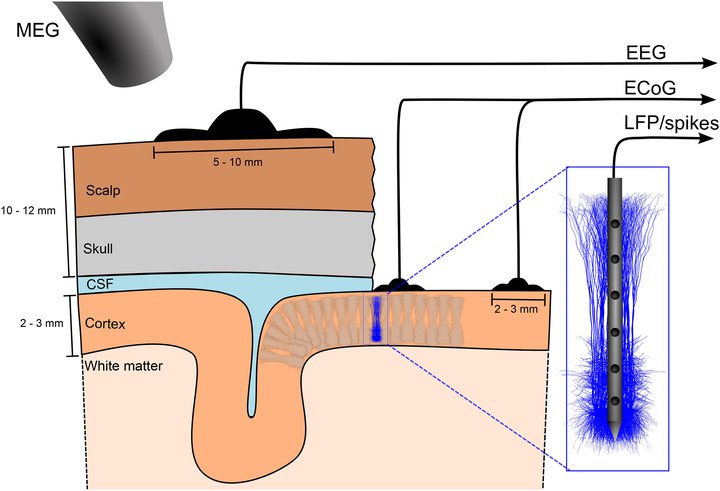
Figure 1: Measures of neural activity in cortical populations: spikes (action potentials) and LFPs from a linear microelectrode inserted into cortical gray matter, ECoG from electrodes positioned on the cortical surface, EEG from electrodes positioned on the scalp, and MEG measuring magnetic fields stemming from brain activity by means of superconducting quantum interference devices (SQUIDs) placed outside the head. For reviews on the biophysical origin and link between neural activity and the signals recorded in the various measurements.
A wide variety of experimental techniques are used in neuroscience today to gain insight into neural function from measured brain signals. But to understand the complex nonlinear dynamics at play in the brain and to explain how the underlying activity gives rise to the signals, computational modeling is required. Simulations provide the crucial link between data generated by experimentalists and these models, a multi-author team, all affiliated with the European Flagship Human Brain Project, now writes in the new article “The scientific case for brain simulation”.
The basis for such simulations has been created on the HBP’s Brain Simulation Platform, a part of the projects Research Infrastructure for brain science that is openly accessible to the neuroscience community. The platform provides a set of continously improved brain network simulators and has driven the construction of computational models and simulations at various scales, from single neurons, to large-scale brain-wide networks.
As the terms can easily be confused, the researchers emphasize that simulation and model are not identical. While mathematical models can embody many different hypotheses about how the brain works in equations and experimental parameters, “brain simulators can be viewed as 'mathematical observatories' to test various candidate hypotheses. A brain simulator is thus a tool, not a hypothesis, and can as such be likened to tools used to image brain structure or brain activity”, the scientists write. Simulation in this context means using sophisticated software tools to set complex models of the brain that represent large numbers of interconnected neurons into motion – and to observe what testable predictions emerge from them.
“The simulation does not represent the goal itself, but serves as a powerful new way for testing competing hypotheses about the brain”, explains Gaute Einevoll, Professor at the Norwegian University of Life Sciences (NMBU) and lead author of the paper. This then serves to enable a systematic “biological imitation game” where models that provide the best predictions of experimental data “win”.
To illustrate the point, Einevoll draws an analogy to the history of physics: "Our project can be compared to Isaac Newton's development of a new branch in mathematics. Newton needed to develop a type of mathematics called calculus to check whether his proposed law of gravitation of how masses such as planets attract each other was correct or not. With it he could calculate the planetary paths in his model and verify that his theory was consistent with observations. With the simulation infrastructure we have developed, we can similarly test whether our candidate network models provide predictions that are consistent with brain measurements. This workflow will be important for further scientific progress, says Einevoll.
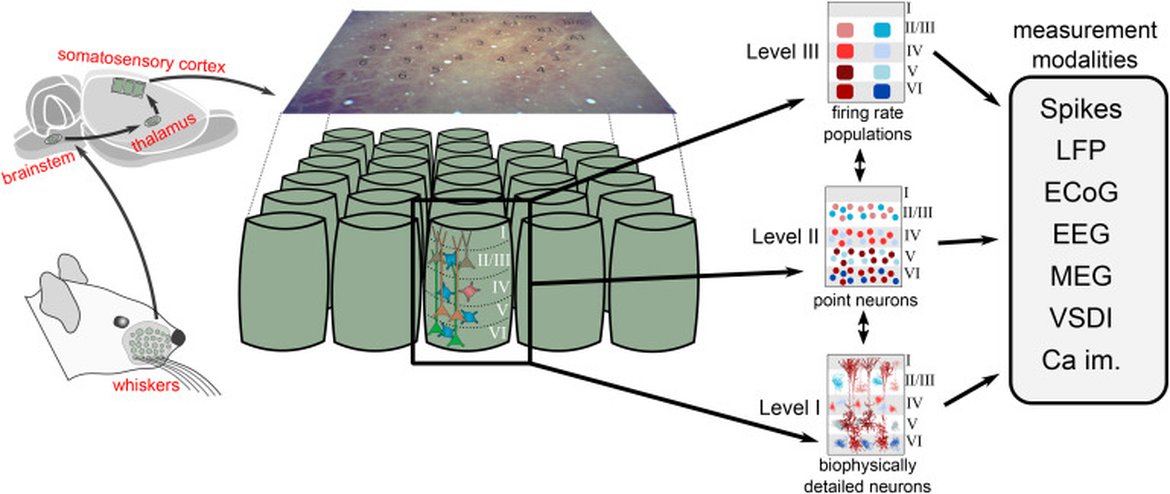
Figure 2: Network dynamics in a cortical column (barrel) processing whisker stimulation in rat somatosensory cortex (left) can be modeled with units at different levels of detail. In the present example, we have a level organization with biophysically detailed neuron models (level I), simplified point-neuron models (level II), and firing-rate models with neuron populations as fundamental units (level III). Regardless of level, the network simulators should aim to predict the contribution of the network activity to all available measurement modalities. In addition to the electric and magnetic measurement modalities illustrated in Figure 1, the models may also predict optical signals—for example, signals from voltage-sensitive dye imaging (VSDI) signals and two-photon calcium imaging (Ca im.).
While the creation of detailed mathematical model has to integrate and generalize a wide range of data provided by experiments, the foundations of the network simulators are simpler and well established, the paper explains – “biophysical principles of how to model electrical activity in neurons and how neurons integrate synaptic inputs from other neurons and generate action potentials. These principles […] are the only hypotheses underlying the construction of brain network simulators. […] This is the reason why many models can be represented in the same simulator and why it is possible to develop generally applicable simulators for network neuroscience.”
“Over the past few years brain network simulators have matured tremendously and so have their scale and applications”, says co-author Markus Diesmann, a computational neuroscientist at Jülich Research Centre and one of the developers of the simulation engine NEST (Neural Simulation Tool). Within the HBP simulation engines like NEURON, Arbor, NEST or The Virtual Brain provide the backbone to address different levels of resolution and biological detail (see below). Each offers specific advantages depending on the question.
"These well tested simulators play an important role in increasing the reproducibility of research by digitized workflows. Making progress here is crucial to be able to build on each other's work", says Sonja Grün, a data analytics expert at Jülich co-authoring the study. “What’s more is that it really creates a new culture of large-scale collaboration across experimental and theoretical neuroscience which we did not have until now”, Diesmann adds.
This collaborative approach, which is well-established in disciplines like physics or astronomy, is a crucial step in approaching the staggering complexity of the brain, the scientists emphasize in their paper:
“Newton said that he had seen further than others because he was ‘standing on the shoulders of giants.’ Likewise, we argue that we need to find a way to stand on the shoulders of each other’s mathematical models to hope to gain a detailed understanding of the functioning of brain networks.”
(ENDS)
(Text in part adapted from HBP partner NMBU https://www.nmbu.no/en/faculty/realtek/news/node/37190)
Contact
Prof. Gaute Einevoll
Norwegian University of Life Sciences
+47-95124536
gaute.einevoll@nmbu.no
Media Contact
Peter Zekert
Human Brain Project
Public Relations Officer
Tel.: +49 (0) 2461 61-96860
Email: p.zekert@fz-juelich.de
Simulation engines in HBP
NEURON and Arbor can run biophysically detailed, multicompartmental neuron models to help understand how dendritic structures affect the integration of synaptic inputs and, consequently, the network dynamics.
More abstracted simulators like NEST can simulate spiking networks of billions of simplified point neurons which model the basic integration of signals and firing of the neurons. This is close to the level of description widely used in AI today, allowing for connections of this type of brain simulations to technology development in this area as well as in robotics. In contrast to most current AI algorithms, NEST and HBP’s neuromorphic hardware systems already capture the fact that real neurons communicate using sparse but robust pulses, and thus operate in a very energy efficient way.
Whole brain-level simulation with the engine The Virtual Brain uses population firing-rate models that summarize dynamics of larger populations of neurons. Such coarse-grained simulation can already help understand and predict large-scale dynamics. Since alterations of such dynamics can be a feature of brain pathologies, like the seizure propagation in Epilepsy, this approach has led to first clinical developments based on patient-specific brain modeling.
The authors

Gaute Einevoll
Professor of Physics at the Norwegian University of Life Sciences and University of Oslo and founder of the Computational Neuroscience Group at the Norwegian University of Life Sciences. He has been a leader or co-leader of the Norwegian national node of the International Neuroinformatics Coordinating Facility (INCF) since 2007. His task in the HBP is to construct biophysical equations and computer simulation tools relating the activity of individual nerve cells to electrical brain signals that, for example, can be measured at the scalp of humans in Electroencephalography (EEG) recordings. (Photo: Håkon Sparre/ NMBU)

Alain Destexhe
Prof. Alain Destexhe, from the Centre National de la Recherche Scientifique (CNRS), France, works on designing realistic models of populations of excitatory and inhibitory neurons, thus linking scales from microscopic (neurons) to large scales (networks or brain areas). He leads the research area of Theoretical Neuroscience in HBP. (Photo: private)

Markus Diesmann
Professor for Computational Neuroscience at RWTH Aachen University; Director, Institute of Neuroscience & Medicine (INM-6, INM-10), Institute for Advanced Simulation (IAS-6), Juelich Research Centre. Research on cortical neuronal network models and simulation technology (NEST) (Photo: Ralf-Uwe Limbach/Forschungszentrum Jülich)

Marc de Kamps
Lecturer in the School of Computing at the University of Leeds working on mean-field methods. These methods allow modellers to create large-scale networks of the brain that retain biological realism without the need to simulate each neuron individually. The aim is to bridge simulations of the neuronal networks underlying cognitive processes, with imaging techniques, which are quite coarse and comprised of the collective signals of many thousands of neurons. (Photo: University of Leeds)

Sonja Grün
Professor for Theoretical Systems Neurobiology at RWTH Aachen University; Director Institute of Neuroscience & Medicine (INM-6, INM-10), Juelich Research Centre. Research on dynamics of neuronal network interactions and data analytics (Elephant)
(Photo: Ralf-Uwe Limbach/Forschungszentrum Jülich)

Torbjørn V. Ness
Researcher in computational neuroscience at the Norwegian University of Life Sciences, with a main focus of calculating experimentally measurable brain signals from simulated neural activity. (Photo: private)

Hans Ekkehard Plesser
Professor of Informatics at the Norwegian University of Life Sciences Visiting scientist, INM-6, FZ Jülich; Deputy Leader of HBP’s High Performance Analytics & Computing Platform. Focus area is NEST Simulation Technology. (Photo: Håkon Sparre/ NMBU)

Viktor Jirsa
Director of Inserm’s Institut de Neurosciences des Systèmes (INS) at Aix-Marseille University and Director of Research at the CNRS. In HBP deputy Leader of the research area of Theoretical Neuroscience and Scientific Leader of the Co-Design Project developing the multimodal and multi-scale HBP human brain atlas. (Photo: private)

Michele Migliore
Senior Research Scientist at the Institute of Biophysics of the Italian National Research Council. His lab is involved in modelling realistic neurons and networks, synaptic integration processes, and plasticity mechanisms. In HBP he is deputy subproject leader of the Brain Simulation Platform. (Photo: private)

Felix Schürmann
Professor at the Blue Brain Project and Brain Mind Institute of Ecole Polytechnique Fédérale de Lausanne (EPFL), Switzerland. In the HBP he is deputy subproject leader of the Brain Simulation Platform and involved in the science and engineering of detailed brain models and respective tools. (Photo: Alain Herzog/EPFL)
About the Human Brain Project
Understanding the organisation of the human brain at all relevant levels is a big challenge, but necessary to improve treatment of brain disorders, create new computing technologies and provide insight into our humanity. Modern ICT brings this within reach. The HBP’s unique strategy uses it to gather, integrate and analyse brain data, understand the healthy and diseased brain, and emulate its computational capabilities. By sharing our tools with researchers worldwide, we aim to catalyse global collaboration.
Unlocking the brain’s secrets promises major scientific, social and economic benefits. One is improved diagnosis and treatment of brain-related diseases; a growing health burden in our ageing population. A second is neuroscience’s potential to contribute to approaches for future ICT, including extreme-scale and neuromorphic computing. The HBP will also contribute to a brain-inspired approach to Artificial Intelligence and robotics.
The HBP studies the brain at different levels, from genomics to higher-level brain functions. To help achieve this goal, the HBP is building an ICT-based research infrastructure to facilitate research collaboration, via the sharing of software tools, data and models. Incorporating inputs from the scientific community, the HBP’s scientists and engineers ensure that our infrastructure meets real research needs. Another aim is to accelerate medical research, by facilitating researchers’ secure access to broader data sets of patient data, as well as HBP tools and models. The HBP also educates young scientists to work across disciplinary boundaries and addresses the ethical implications of its work. Finally, it helps to integrate global brain research efforts and leads Europe’s contribution in this field.



