MRI and AI to develop a brain virtual biopsy tool
20 May 2019
A novel tool has been developed to meet the needs of both fundamental research on the human brain, in particular to decode the cyto- and fiber architectures of the cerebral cortex in vivo, and the clinical research to provide clinicians with a virtual biopsy tool which could eventually replace invasive surgical biopsies.
In the frame of the European Flagship Human Brain Project, Cyril Poupon and Kévin Ginsburger from Neurospin (Frédéric Joliot Institute for Life Sciences, CEA Paris-Saclay, France) and their research partners Markus Axer and Felix Matuschke from Research Centre Juelich (Institute of Neuroscience and Medicine INM1, Juelich, Germany) are developing a novel computational approach based on large scale numerical simulations and machine learning algorithms to decode healthy and pathological brain tissue cellular organization and axonal architecture using diffusion MRI and Polarized Light Imaging (PLI) modalities.
The decoding tool relies on a simulation pipeline which requires High Performance Computing facilities. Indeed, the diffusion MRI or PLI signal is synthesized from a huge collection of ultra-realistic brain tissue virtual samples, using massively parallel simulation codes. These virtual samples are created using a novel generative method recently published in the journal NeuroImage.
 The study has been featured on the cover of the journal (June 2019, vol. 193)
The study has been featured on the cover of the journal (June 2019, vol. 193)
In the article, the MEDUSA (Microstructure Environment Designer with Unified Sphere Atoms) software is presented, which allows constructing realistic cellular environments from any brain area. MEDUSA enables to create in silico brain tissue samples with unprecedented realism, using a reduced number of input parameters.
This novel tool can be used to simulate the diffusion MRI or PLI signal from any region of the brain, thus enabling to learn the “signature” of each specific brain tissue configuration using artificial intelligence algorithms. It has been developed to meet the needs of both fundamental research on the human brain, in particular to decode the cyto- and fiber architectures of the cerebral cortex in vivo, and the clinical research to provide clinicians with a virtual biopsy tool which could eventually replace invasive surgical biopsies.
The MEDUSA software is already being employed in a first simulation campaign at the CEA Very Large Computing Centre (CCRT, Bruyères-le-Châtel), which aims at training algorithms to quantify the degree of swelling (also called beading) of axons after ischemic strokes. A larger simulation campaign for which a computational resource application has been sent to the GENCI (Grand Équipement National de Calcul Intensif) is also under preparation to map the brain white matter microstructure in vivo.
(adapted with permission from: http://joliot.cea.fr/drf/joliot/Pages/Actualites/Scientifiques/2019/IRM-et-IA-pour-developper-outil-biopsie-virtuelle-cerveau.aspx)
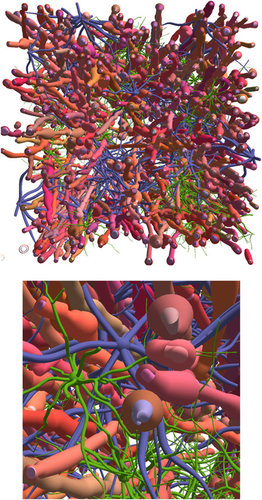
Virtual brain white matter sample (100 µm on each side of the cube) constructed with the MEDUSA software, including 200 myelinated axons, and numerous astrocytes and oligodendrocytes. © K. Ginsburger / C. Poupon / CEA
Original Publication:
Ginsburger, K., Matuschke, F., Poupon, F., Mangin, J. F., Axer, M., & Poupon, C. (2019). MEDUSA: A GPU-based tool to create realistic phantoms of the brain microstructure using tiny spheres. https://doi.org/10.1016/j.neuroimage.2019.02.055



