EBRAINS Infrastructure Training on
MODEL VALIDATION
4–7 May 2021 | Virtual Event
Background:
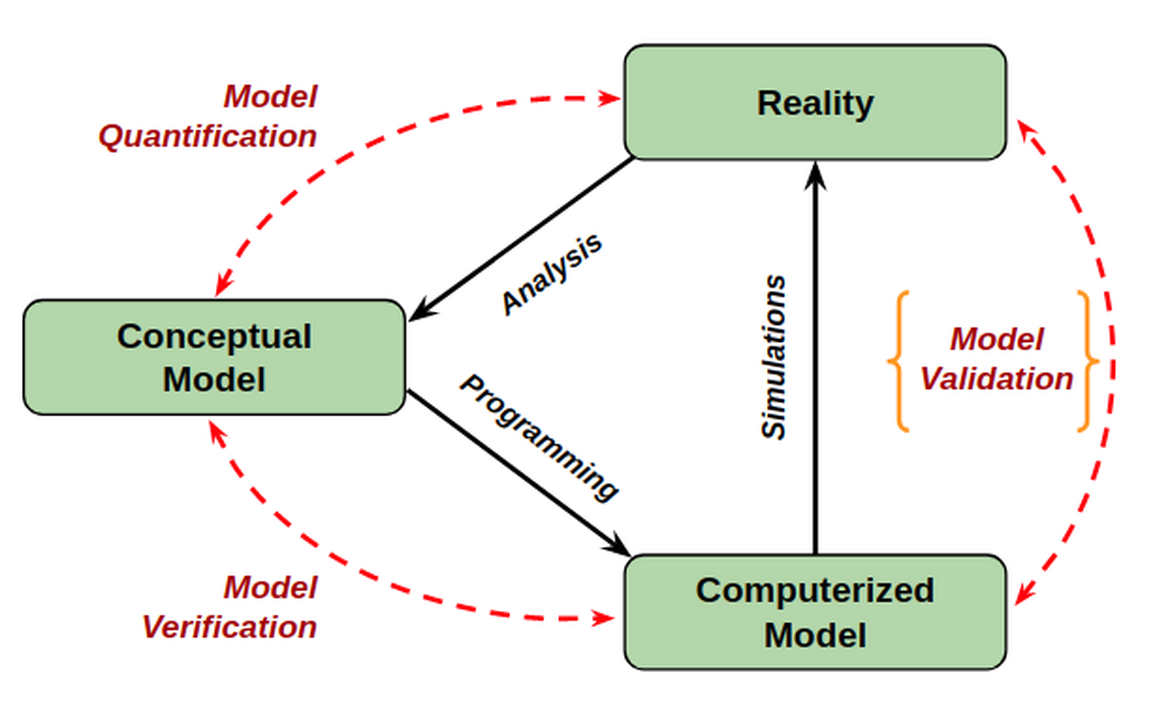
A major shortcoming in computational neuroscience in the absence of a well-defined system for the validation and comparison of models. This impedes progress, with modellers often working in isolation from experimentalists, and leads to unecessary difficulty in comparing published models to each other or to new data. The process of model validation is typically manual, qualitative, and limited in the number of properties tested. Explicitly quantifying the differences between simulation and experimental results, decoupling test definitions from individual models, and automating the processes of finding and running tests, can greatly increase the productivity of the simulation-validation cycle, thereby reinforcing the scientific method.
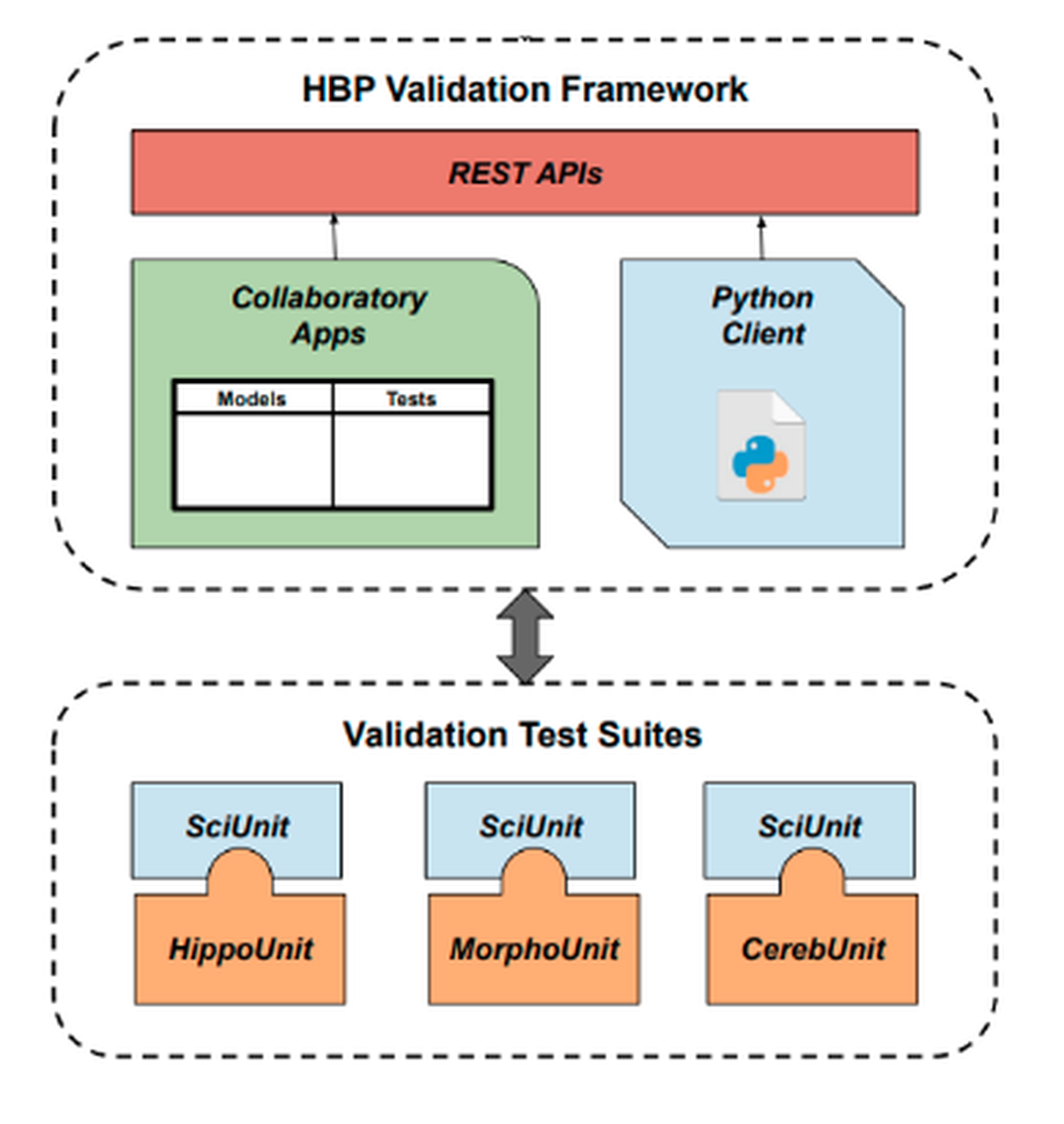
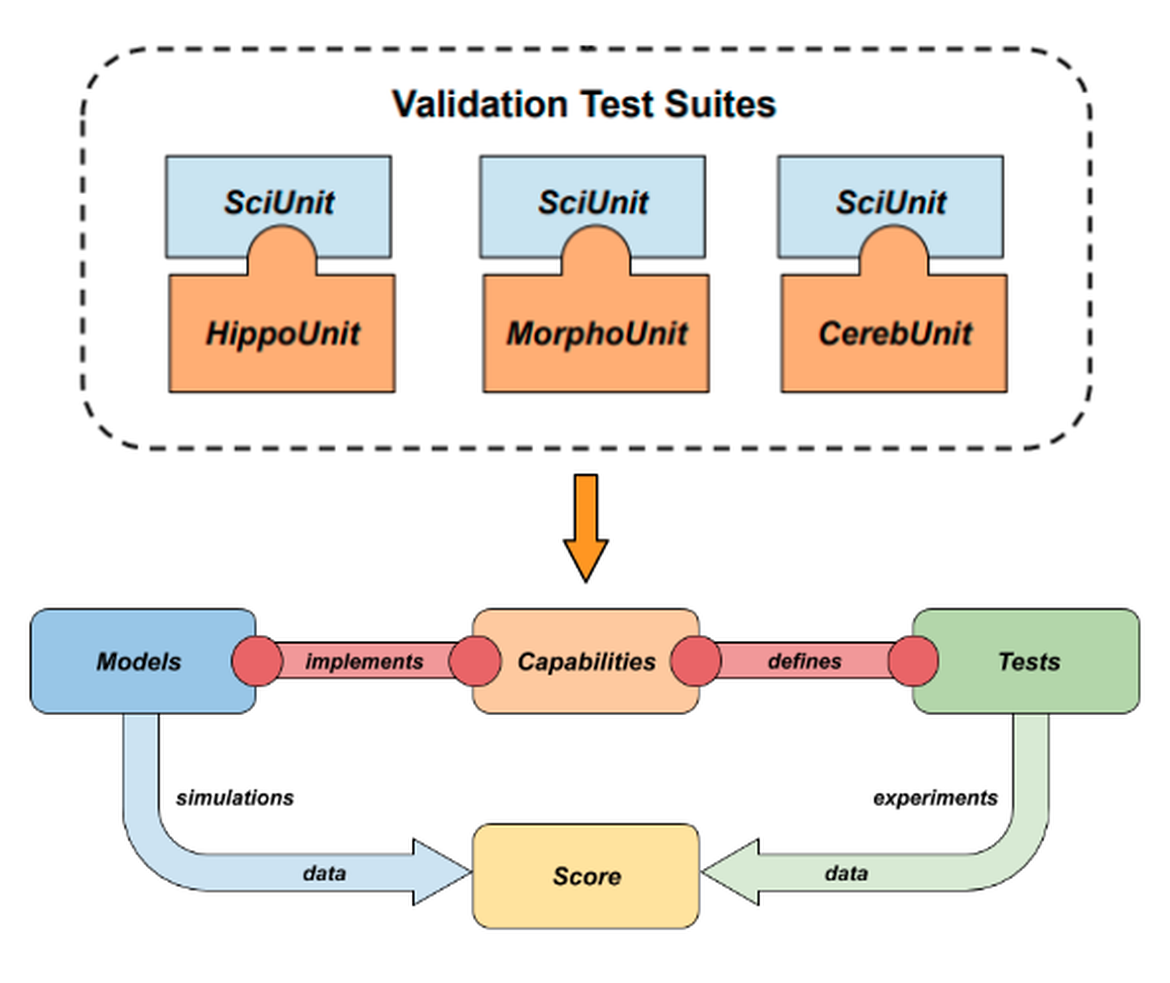
The EBRAINS Model Validation Framework provides web-based tools for working with computational models, and the validation of such models against experimental data with a systematic, unbiased and reproducible approach. It consists of a REST web service and two clients: the Model Catalog collaboratory app and a Python client. Underlying metadata are stored in the Knowledge Graph, thereby coupling it with other EBRAINS workflows. The framework broadly allows users to (i) create, edit, search and view metadata about models and validation tests, and (ii) register, search, view and compare the results of validations run on models.
This training will guide modellers and experimentalists in model validation (comparing simulation results to experimental data) using the EBRAINS Model Validation Framework.
Participants will learn how to:
- develop model-agnostic validation tests
- adapt existing models so they can be more easily validated
- use the EBRAINS model and test catalogue
- register, search, view and compare the results of validation tests
The training takes place over four days, with two days of presentations and hands-on demos, followed by two days of participants working on their own validation projects with the assistance of the tutors.
PROGRAMME Up-to-date online version of the Programme!
The training is open to all interested researchers working with brain models. The hands-on sessions and project work (last two days) would be undertaken using Python programming language. Some familiarity with Python would be useful, but not essential.
For this 2nd part of the training, you are welcome to send us a short abstract, describing your model.
Abstracts should include:
- A short description of your model
- Simulators/tools used for developing and simulating models
- Overview of validation(s) you wish to implement during the training (specify at least one validation test), along with target (experimental) reference data
- If you have already some questions or comments, you are welcome to mention them
- If your model is already published, please include references at the end of the abstract
Please send your abstract latest until 28 April 2021, 5pm (CET) to training-support@humanbrainproject.eu.
REGISTRATION
CONFIRMED SPEAKERS AND TUTORS
 Dr. Shailesh Appukuttan is a postdoctoral researcher in the Department of Integrative and Computational Neuroscience of the Paris-Saclay Institute of Neuroscience (CNRS, Université Paris-Saclay), France, with the Neuroinformatics research group. He is interested in the application of computational techniques to further biological research. As part of the Human Brain Project (HBP), his work involves the design and development of a model validation framework for neuroscience, and its integration into existing model development workflows, along with development of other related scientific tools.
Dr. Shailesh Appukuttan is a postdoctoral researcher in the Department of Integrative and Computational Neuroscience of the Paris-Saclay Institute of Neuroscience (CNRS, Université Paris-Saclay), France, with the Neuroinformatics research group. He is interested in the application of computational techniques to further biological research. As part of the Human Brain Project (HBP), his work involves the design and development of a model validation framework for neuroscience, and its integration into existing model development workflows, along with development of other related scientific tools.
Personal webpage: http://shailesh-appukuttan.scienceontheweb.net/

Dr. Andrew Davison is a senior research scientist in the Department of Integrative and Computational Neuroscience of the Paris-Saclay Institute of Neuroscience (CNRS, Université Paris-Saclay), France, where he leads the Neuroinformatics research group. His research interests focus on biologically detailed modelling of neuronal networks (particularly the mammalian visual system), and on developing tools to improve the reliability, reproducibility, and efficiency of biologically realistic simulations.
 Dr. Pedro Garcia-Rodriguez is currently a Postdoc in Andrew P. Davison's team of the Neuroscience Paris-Saclay Institute. He completed a PhD in Computational Neuroscience at Pompeu Fabra University, Barcelona, Spain in 2012. Later on, he got involved in the statistical validation of biophysically detailed models for Basal Ganglia and Hippocampus brain regions, as part of the HBP project. His technical background includes computer programming, neuroscience simulators or APIs such as NEST, NEURON and PyNN, physics, non-linear dynamical systems and stochastic processes theory.
Dr. Pedro Garcia-Rodriguez is currently a Postdoc in Andrew P. Davison's team of the Neuroscience Paris-Saclay Institute. He completed a PhD in Computational Neuroscience at Pompeu Fabra University, Barcelona, Spain in 2012. Later on, he got involved in the statistical validation of biophysically detailed models for Basal Ganglia and Hippocampus brain regions, as part of the HBP project. His technical background includes computer programming, neuroscience simulators or APIs such as NEST, NEURON and PyNN, physics, non-linear dynamical systems and stochastic processes theory.
 Prof. Rick Gerkin researches the design and validation of neural models from single neurons to human psychophysics. As the lead developer of SciUnit he is interested in a Pythonic approach to introducing test-driven development into the modeling process. Other efforts include the curation of diverse datasets from which such model validation tests can be constructed. He is also the Data Science Core lead on the "Cracking the Olfactory Code" consortium sponsored by US NIH, integrating physiological, optical, genomic, and behavioral data into a unified analysis and discovery framework.
Prof. Rick Gerkin researches the design and validation of neural models from single neurons to human psychophysics. As the lead developer of SciUnit he is interested in a Pythonic approach to introducing test-driven development into the modeling process. Other efforts include the curation of diverse datasets from which such model validation tests can be constructed. He is also the Data Science Core lead on the "Cracking the Olfactory Code" consortium sponsored by US NIH, integrating physiological, optical, genomic, and behavioral data into a unified analysis and discovery framework.
 Robin Gutzen is a PhD student at the Institute for Computational and Systems Neuroscience at the Research Center in Jülich, Germany. His research focuses on the statistical analysis and comparison of neural population dynamics in the form of spiking activity and field potentials. Beyond that, he is interested in the development of scientific software tools to facilitate, for example, validation testing and the integration of heterogeneous data into analysis workflows. Personal Webpage: https://rgutzen.github.io/
Robin Gutzen is a PhD student at the Institute for Computational and Systems Neuroscience at the Research Center in Jülich, Germany. His research focuses on the statistical analysis and comparison of neural population dynamics in the form of spiking activity and field potentials. Beyond that, he is interested in the development of scientific software tools to facilitate, for example, validation testing and the integration of heterogeneous data into analysis workflows. Personal Webpage: https://rgutzen.github.io/
 Sára Sáray is a doctoral candidate in the Department of Cerebral Cortex Research of the Institute of Experimental Medicine and the Faculty of Information Technology and Bionics, Pázmány Péter Catholic University, Hungary. She is interested in the systematic construction and validation of detailed models of hippocampal neurons based on electrophysiological data.
Sára Sáray is a doctoral candidate in the Department of Cerebral Cortex Research of the Institute of Experimental Medicine and the Faculty of Information Technology and Bionics, Pázmány Péter Catholic University, Hungary. She is interested in the systematic construction and validation of detailed models of hippocampal neurons based on electrophysiological data.
Her research interests include the computational modeling of dendritic integration in hippocampal pyramidal cells.
-
Model Catalog (Validation Framework) App
https://model-catalog.brainsimulation.eu/
- Model Catalog (Validation Framework) App - Docs:
https://model-catalog.brainsimulation.eu/docs/ - Validation Framework REST APIs:
https://validation-v2.brainsimulation.eu/docs - Validation Framework Python Client Docs:
https://hbp-validation-client.readthedocs.io/ - SciUnit Tutorials:
https://sciunit.readthedocs.io/en/latest/quick_tutorial.html - HippoUnit Repository:
https://github.com/KaliLab/hippounit - HippoUnit Documentation:
https://hippounit.readthedocs.io/
FAQs & all you need to know
Please make sure to register with your institutional email address.
The link to our virtual conference platform as well as the online credentials will be sent out to all registered participants a few days prior to the event.
To make this virtual training an interactive experience, it is important that you have access to a stable internet connection, good audio (with microphone) and ideally (not mandatory) a webcam for video communication.
Also, to have the full conference experience it is important that you have installed the Zoom Desktop Client application (v5.3.0 or higher) and that you access the conference platform via the latest version of Google Chrome.
If you have any questions about the event, please contact us at training-support@humanbrainproject.eu
All times in the programme schedule are UTC+2 = CEST.
To prevent missed sessions, we recommend to use a time zone converter in advance.