SENSEI
SENSEI brings together teams with expertise in signal and image processing (Research Center “E. Piaggio”, University of Pisa, Italy), basic neuroscience, live and super-resolution imaging analysis (INSERM126-IPNP, Imaging facility of the Center for Psychiatry and Neuroscience Paris, France) and development of new imaging modalities for high-content and super-resolution microscopy (Lab for Nanobiology, Department of Chemistry, University of Leuven, belgium), working in synergy for advancing breakthroughs in 3D neuronal segmentation and morphometrics.

Project description
Digitizing a complete and high-fidelity map of the neurons populating the mammalian brain, comprising spines and axons, is a central endeavour for neuroscience research. In fact, our ignorance of the brain micro-circuitry underpins the difficulty of deeply exploring the function of complex neural circuits and understanding fundamental (patho)physiological brain processes. While marked improvements in imaging tools (e.g., 3D microscopy techniques) and protocols (e.g., membrane probes and clarification protocols) have enabled the visualization of large brain volumes at cellular and sub-cellular resolution, the state of the art continues to be principally limited by the difficulties in neuron segmentation, which is still far from ground-truth. This is mainly due to the lack of image processing algorithms able to deal with 3D microscopic data acquired from different imaging modalities, representing specimen processed with different procedures and belonging to different animal species.
SENSEI’s objectives are to accurately quantify neuronal morphology at tissue and molecular levels through the development of intelligent segmentation-based image processing algorithms and to improve the quality of neuronal imaging using new membrane probes for conventional and emerging super-resolution imaging technologies. Specifically, we will develop an algorithm for isolating single neurons or smaller neuronal structures from 3D datasets representing the intricacies of the brain micro-structure.
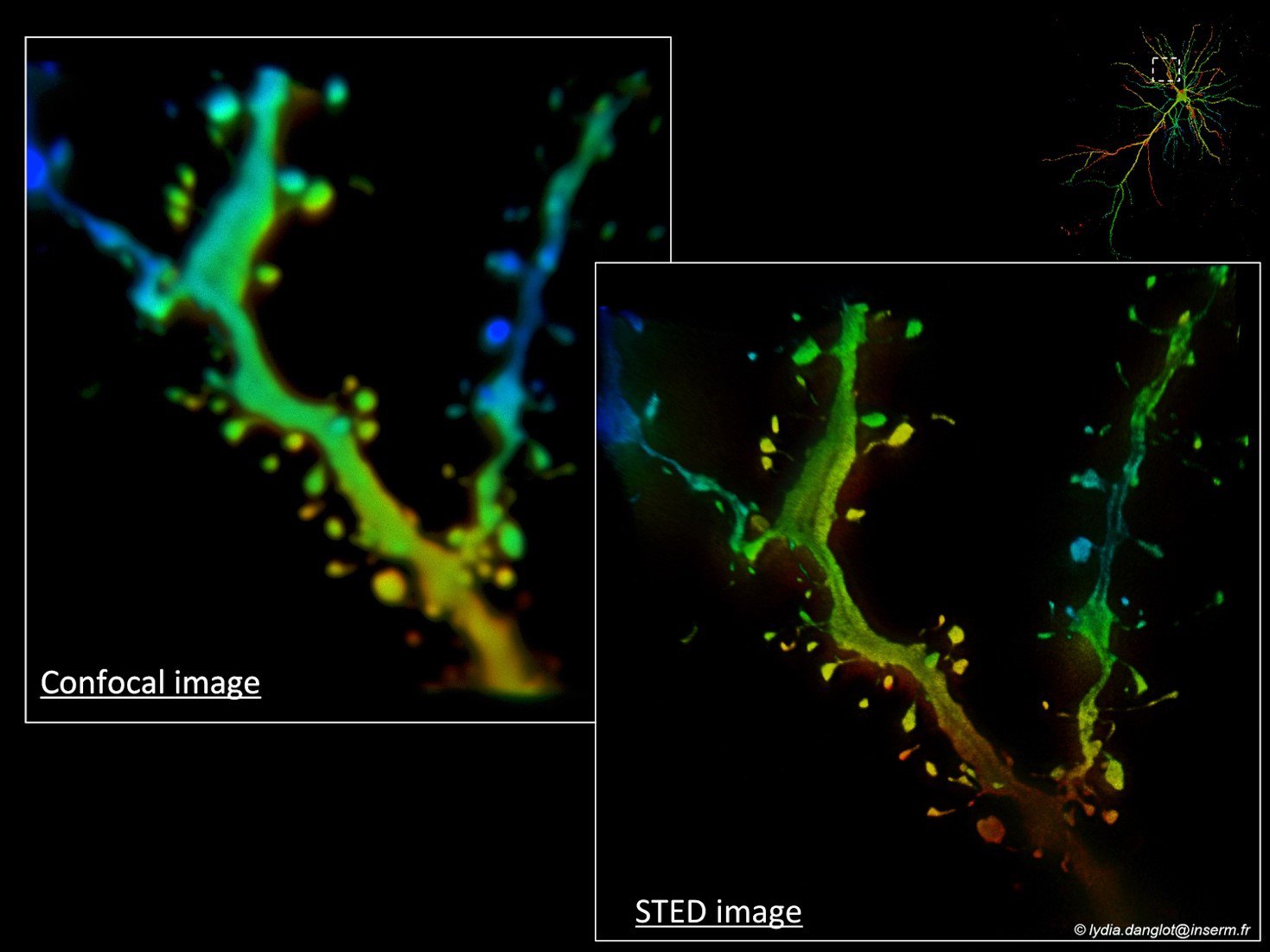
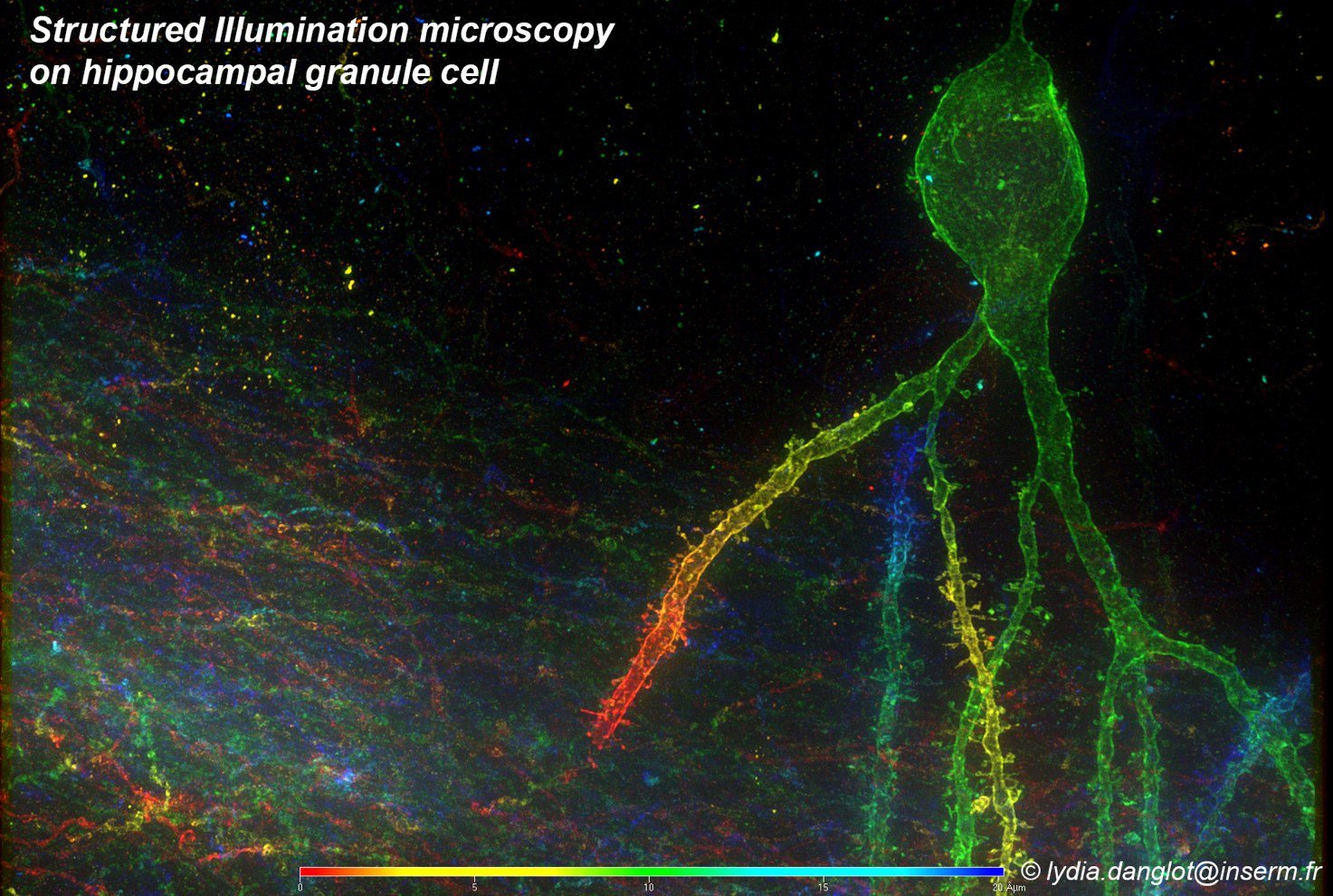
This project will study several mammalian species (including human) both in vitro for 2D (hippocampal, cortical neurons) and in situ for 3D (tissue slices and clarified brains) in cortex, cerebellum and hippocampus. New imaging modalities and segmentation protocols will be tested on human brain tissues coming from surgical resections. Samples will be imaged both with confocal microscopy, accounting for 80 % of user needs, and advanced imaging modalities, thus bringing either high resolution details on fixed dendritic spines (e.g., STED, STORM) or on live neurons (i.e. 3D SOFI). In addition to the sub-diffraction resolution, an optical device encoding probe information in the microscope point-spread function will be constructed, allowing the fast acquisition of cellular nanostructuring in full-3D.
SENSEI aims at facilitating 3D reconstruction of neuronal morphology and circuitry, aiding neuro-anatomical mapping, and generating models which can be used for making predictions about higher-level brain organization. The detailed analyses of neurons might shed new light on the normal development of dendritic and axonal arbours and on how these processes are altered in neuro-pathologies.
The main objective of SENSEI is to deliver a comprehensive breakthrough solution for neuronal imaging and segmentation using a robust and accurate algorithm for reconstructing 3D neuronal morphology based on datasets acquired using advanced and emerging tools.
This goal will be achieved by addressing the following specific objectives:
- Testing of new membrane probes and improving image acquisition of neuronal cells in 2D cultures and in 3D tissues with conventional and super-resolution microscopy;
- Developing a framework for the statistical characterization of microscopic images;
- Improving fast imaging of live and fixed neurons using super-resolution 3D SOFI imaging;
- Improving a neural segmentation algorithm based on statistical models of image noise and cell signal and testing it using samples from different neuronal cells, brain sites and animal species, from rodents up to humans;
- Extending the algorithm for the application at different scales (super-resolution and live imaging);
SENSEI aspires to the HBP Flagship directives, which emphasize the importance of the development of tools for high-fidelity 3D reconstruction of the intricacies of the mammalian brains, through a collaborative, trans-disciplinary environment.
SENSEI outcomes will provide a set of tools for accurately reconstructing brain micro-circuitry and for extracting quantitative morphometric information of neuron shape, size and complexity. The tools delivered in SENSEI could be used for monitoring neuronal morphological abnormalities associated with neuropathies and studying neuronal development, as well as for highlighting any differences with the human counterpart.
Another possible integration between HBP and SENSEI regards the study of human brain organization. In fact, SENSEI aims at developing tools and frameworks that can facilitate the data processing for mapping brain organization, and in particular human hippocampal complex, at the micro- and nano-scale.
SENSEI aims at providing 3D reconstructions of different neuron types in their 3D arrangement while preserving the neuron volumetric information. Future developments could regard, the enrichment of segmented neurons by SENSEI with the information about neuron orientation and position within the brain, aiding at building the Rodent and Human Atlases. Moreover, the volumetric information provided by our tool for single-cell isolation from 3D microscopic datasets will provide valuable inputs to the models developed in the Brain Simulation Platform, i.e. allowing the simulation of electrophysiological cell behaviour.
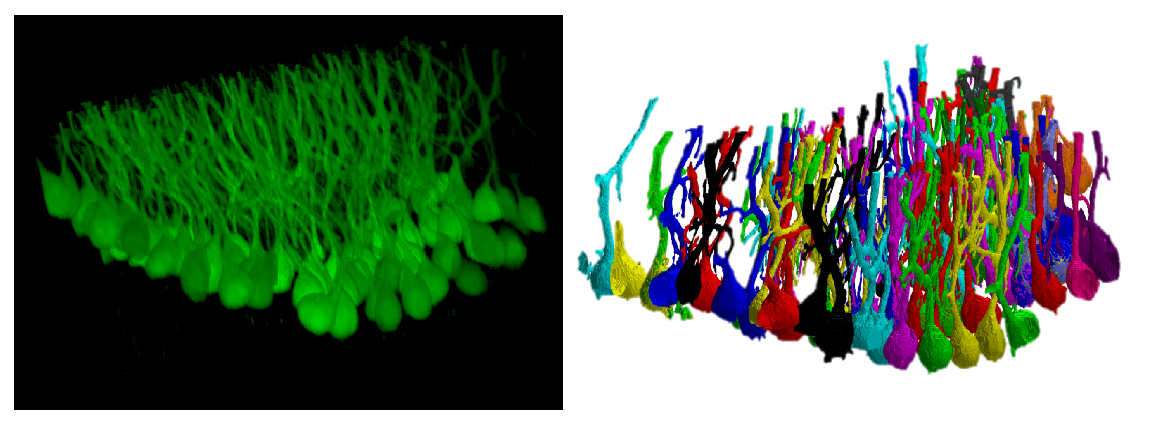 A confocal dataset representing Purkinje cells from clarified cerebellar tissue and the segmentation results.
A confocal dataset representing Purkinje cells from clarified cerebellar tissue and the segmentation results.
Partnering Organisations






Biography

Nicola Vanello (Project Coordinator)
Associate Professor, member of the Research Center “E. Piaggio” and with the Dipartimento di Ingegneria del’Informazione. His research activity is related to models and methods for biomedical signal and image processing. Specifically, his skills concern the development of exploratory and confirmatory models for data analysis using optical imaging, electroencephalography and magnetic resonance imaging (MRI). His research interests cover the development of methods for confocal microscopy image processing.

Lydia Danglot is a neurobiologist at the Institute of Psychiatry and Neuroscience of Paris and permanent Inserm researcher trained at the Ecole Normale Superieure (Antoine Triller’s lab.) and Pierre et Marie Curie University. Her research focuses on neuronal morphogenesis of the synapse during learning and memory in normal and pathological context (Alzheimer disease). She has a strong involvement in Advanced Microscopy Technics (SIM, STED, STORM, Nature Comm, 2018) and in the development of new fluorescent dyes for Neuroscience (JACS 2018, Cell chemical Biology 2019). She’s Board member of the National Research Group ImaBio gathering more than 100 labs and facilities on advanced microscopy for biology. She ‘s part of the organizing committee of the French Microscopy School Mifobio and Inserm international workshop. She’s national coordinator of the France Bio Imaging infra-structure users committee.

Dr. Peter Dedecker Assistant Professor at KUL, heads the lab for Nanobiology at the KU Leuven. His research focuses on the live visualization of cellular regulation and processes at the nanoscale level, by combining the development of ‘smart’ fluorophores with custom imaging solutions and instruments. Hi research group is funded independently, with grants from the Flemish FWO, Horizons2020, the Hercules Foundation, Marie Curie postdoctoral fellowship, and an ERC starting grant. In addition to extensive facilities for fluorophore development and characterization, the group also possesses advanced imaging instruments and has access to the full range of imaging facilities available within the fluorescence center of excellence at the Faculty of Sciences.

PhD in Bioengineering and a BSc in Physics. Director of Research Center “E. Piaggio” and Director of the Centro 3R (Interuniversity Center for Promoting the 3Rs Principles), Professor of Bioengineering at the Department of Information Engineering. She also co-heads the NanoBioscopy Lab (a state of the art imaging facility at UNIPI with confocal, multi-photon microscopy, optical coherence and microtomography). She is a member of the Italian Ministry of Health’s expert group on the implementation of the 3RS. She has coordinated and participated in several FP7 and H2020 projects.

Francesca Vaglini
PhD in Clinical Neuropharmacology. Researcher at the Department of Translational Research and New Technologies in Medicine and Surgery. Her research mainly focuses on animal models of neurologic disease, in particular she contributed to develop and characterize toxicant-induced models for Parkinson's disease, including MPTP and rotenone-induced dopaminergic cell death and she studied a transgenic model of Autism Spectrum Disorders.

PhD in Automation, Robotics and Bioengineering. For the high quality of her work, she won the “Massimo Grattarola” award for the best Italian thesis in the field of neuroinformatics and neuroengineering. She is actually a Post-Doc Researcher at the Research Center “E. Piaggio” thanks to a Fellowship of Fondazione Umberto Veronesi, an important Italian institution in the field of oncology and neuroscience. Her work mainly focuses on customizing clarification and acquisition procedures as well as developing open-source algorithms for both single neuron segmentation and 3D morphometric extraction from 3D images of brain tissue.

Alejandro Luis Callara is a postdoctoral researcher at the Research Center ”E. Piaggio” at the University of Pisa. He received his bachelor’s Degree in Biomedical Engineering in 2012 and his master’s Degree in Biomedical Engineering in 2015 from the University of Pisa, Italy. He received his PhD in Information Engineering in 2019 from the Dipartimento di Ingegneria dell’Informazione, University of Pisa. His main research activity is related to models, methods and pipelines for biomedical signal and image processing. He is particularly skilled with electroencephalography (EEG) and functional magnetic resonance imaging (fMRI), and with methods for inferring brain connectivity from these kind of data. His research interests include the development of models, algorithms and tools for the segmentation and 3D morphometric analysis of single neurons from microscopy data.
Publications
Callara, A.L., Magliaro, C., Ahluwalia, A., Vanello, N. (2020) “A Smart Region-Growing Algorithm for Single-Neuron Segmentation From Confocal and 2-Photon Datasets” Frontiers in Neuroinformatics, 14, art. no. 9
C. Magliaro, A.L. Callara, G. Mattei, M. Morcinelli, C. Viaggi, F. Vaglini, A. Ahluwalia, Clarifying CLARITY: Quantitative Optimization of the Diffusion Based Delipidation Protocol for Genetically Labeled Tissue, Front Neurosci. 10 (2016).
C. Magliaro, A.L. Callara, N. Vanello, A. Ahluwalia, A Manual Segmentation Tool for Three-Dimensional Neuron Datasets, Front Neuroinform. 11 (2017) 36. doi:10.3389/fninf.2017.00036.
Collot M*, Ashokkumar P, Anton H, Boutant E, Faklaris O, Galli T, Mély Y, Danglot L*, Klymchenko AS*. MemBright: A Family of Fluorescent Membrane Probes for Advanced Cellular Imaging and Neuroscience. Cell Chemical Biology. 2019 Apr 18;26(4):600-614.e7. Acta Neuropathol. 2018 Jun;135(6):839-854. * corresponding authors
Lagache T, Grassart A, Dallongeville S, Faklaris O, Sauvonnet N, Dufour A, Danglot L*, Olivo-Marin JC*.Mapping molecular assemblies with fluorescence microscopy and object-based spatial statistics. Nature Comm. 2018 Feb 15;9(1):698. * corresponding authors
Androuin A, Potier B, Nägerl UV, Cattaert D, Danglot L, Thierry M, Youssef I, Triller A, Duyckaerts C, El Hachimi KH, Dutar P, Delatour B, Marty S. Evidence for altered dendritic spine compartmentalization in Alzheimer's disease and functional effects in a mouse model. Acta Neuropathol. 2018 Jun;135(6):839-854.
Vandenberg W., Leutenegger M., Duwé S, Dedecker P. An extended quantitative model for super-resolution optical fluctuation imaging (SOFI). Optics Express, 2019, September, 27.: 25749 - 25766.
De Zitter E., Thédie D., Mönkemöller V., Hugelier S., Beaudouin J., Adam V., Byrdin M., Van Meervelt L., Dedecker P., Bourgeois D. Mechanistic investigation of mEos4b reveals a strategy to reduce track interruptions in sptPALM. Nature Methods, 2019, July, 16: 707–710
Key facts
Time frame: 2020 - 2023
Origin: FLAG-ERA JTC2019



