Access to the state-of-the-art facilities and resources of HBP Partner institutions, to foster collaboration and carry out cutting-edge scientific research.
HBP and HBP Partners provided access to three types of Facility Hubs during the last phase of the HBP:
- Physical facilities (e.g. microscopes)
- Competence centres providing specialised software support going beyond the EBRAINS High Level Support Team
- The European Institute for Theoretical Neuroscience (EITN)
Facility Hubs were aligned with the Fenix supercomputing network to provide user access to computing resources, e.g. for data storage, analysis, and simulation.
The Facility Hubs were contributions from HBP Partners to the new EBRAINS research infrastructure, and complemented the EBRAINS services on offer.
Access and Terms of Use
Users from academia and industry world-wide were eligible to apply for access to HBP Facility Hubs.
How access was organised
Interested users of the Facility Hubs were asked to contact the respective Facility Hub contact for a first discussion on the potential project and support as well as the feasibility. Users must bring their own funding for the project which goes beyond the offer/support by the Facility Hub. Facility Hub support could also be included in applications to Calls.
Users were asked to submit a short project description to the Facility Hub contact using the template provided by the Facility Hub contact, which included a project summary, complementarities and synergies with the Human Brain project, and the expected outputs. Since most of the Facility Hubs required extensive compute resources (e.g. for simulations or data storage), the Facility Hubs were aligned with the Fenix research infrastructure. Facility Hubs had applied via ICEI (Interactive Computing E-Infrastructure for the Human Brain Project) for a certain amount of compute and data resources which they could make available via "wide access" to users of the hub. If users needed more resources than supplied at the Hubs, they had to submit a request via ICEI.
The respective Facility Hub decided which projects were supported according to its rules and regulations. Criteria for the selection were scientific excellence, impact and contribution to the aims of the Human Brain Project, EBRAINS and the Facility Hub, as well as ethical compliance with the regulations of the Hub.
Before the project started, a Usage Agreement was signed. The usage needed to be in agreement with the EBRAINS General Terms of Use and the EBRAINS Access Policy as well as with the Facility Hub-specific licence and rules. Results (e.g. datasets, software, models) needed to be curated in EBRAINS and publications or other communications using results of the project needed to acknowledge the Human Brain Project and EBRAINS (and the ICEI project, if any ICEI resources were used).
Reporting:
Users were asked to deliver a short project report by the end of the project according to the template which was provided by the Facility Hub contact before the project start.
Offers after September 2023
Physical facilities
This section describes the offers of the Facility Hubs at the end of the HBP in September 2023. The facilities might have changed or been updated since then.
Two-photon imaging and ensemble recordings, both conducted in behaving, task-performing animal models, are illuminating our understanding of how cognitive, perceptual and motor functions originate from the behaviour of large, cooperating populations of neurons. The University of Amsterdam (UvA) facility hub and competence centre offers access to a Leica fast resonant two-photon scanner and high-density ensemble recording setups using Neuropixels and other silicon probe configurations. The core expertise in electrophysiological, optical and behavioural recordings is supplemented by opto- and chemogenetics, large-scale data analysis centred on population decoding and computational modelling.
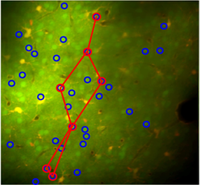
The UvA can host medium (2-6 mo.) or longer (>6 mo.) term projects for PhD students, postdocs and research staff who need to acquire multiple technical skills and/or want to acquire data for a project. Shorter or remote visits are possible for researchers wanting to learn individual skills related to data-analysis. Organising online and on-site courses and webinars for large groups is also possible.
The UvA harbours one of the largest groups for two-photon in vivo neuronal imaging and ensemble recordings in Europe, which is uniquely distinguished by its development of sophisticated behavioural paradigms testing multisensory perception, memory and consciousness, a high level of multidisciplinary integration and technical versatility.
Additional information: Systems and Neuroscience Group web page
The hub “3D-Polarised Light Imaging” at the Institute of Neuroscience and Medicine (INM-1), Forschungszentrum Jülich, Germany, offers hands-on insights into the research of the brain’s fibre architecture at microscopic scales, based on unstained histological brain sections.

The imaging technique of choice is referred to as 3D-Polarised Light Imaging (3D-PLI) and has been uniquely developed at the INM-1 over the last ten years. It basically utilises the optical birefringence of nervous tissue, mainly induced by myelinated axons. 3D-PLI makes studies of the fibre architecture (e.g. identification and tracing of fibre pathways and tracts, and quantification of fibre orientations and myelin distributions) in 50μm thick brain sections possible.
The hub offers possibilities
(i) for initial consultation about the topic of fibre-targeted microscopy,
(ii) to carry out a feasibility study, or
(iii) to obtain independent cross-validation on a small scale
for three weeks per year.
An approved project exclusively includes introductory training and guidance in brain cryo-sectioning and tissue mounting specifically required for 3D-PLI applications. Section scanning will be closely supervised by an on-site expert and carried out using one of the available polarising microscopes. Finally, comprehensive image analysis will be performed utilising the FENIX infrastructure, including high-performance computing, data storage and sharing resources at the nearby Jülich Supercomputing Centre.
Additional Information: Fibre Architecture and Polarized Light Imaging website
The combination of focused ion beam (FIB) milling and scanning electron microscopy (SEM) is a method that permits the rapid and automatic serial reconstruction of large tissue volumes. The FIB/SEM facility —located at the Centro de Tecnología Biomédica (Laboratorio Cajal de Circuitos Corticales, UPM/CSIC), Universidad Politécnica de Madrid )— is intended to be a leading center for (i) detailed synapse studies using 3D electron microscopy and (ii) generation of high-resolution images of brain regions in the millimeter to nanometer range.
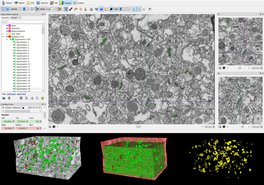
For detailed information see ‘FIB/SEM: Description and applications to study the brain’ at http://cajalbbp.es/CBB
This FIB/SEM facility is associated with high-level scientific and technician staff who will provide the necessary expertise in electron microscopy to ensure that any scientist can benefit from this facility, regardless of whether or not they have experience in electron microscopy. The profiles involved in the projects will be Senior Researchers, High-Level Technicians and Laboratory Technicians. The number of people involved will be based on the project needs, with this experienced staff devoting 20% of their time to supporting the selected projects.
The UPM can host medium (2–6 mo.) or longer (>6 mo.) term projects for PhD students, postdocs and research staff who need to acquire multiple technical skills and/or want to acquire data for a project.
Light-Sheet Fluorescence Microscopy (LSFM) of cleared brain samples offers the unique opportunity of exploring brain complexity on large scales yet at cellular resolution. The European Laboratory for Non-Linear Spectroscopy (LENS) has gathered long-standing experience in the whole LSFM pipeline, comprising sample preparation (clearing and staining), the actual imaging using custom-designed optical setups, and image post-processing (volumetric stitching, compression, feature segmentation). The LENS facility hub offers access to two microscopy setups capable of high-speed and high-resolution imaging with simultaneous two-colour acquisition. One system can accommodate large bulky samples such as a whole mouse brain, while the other microscope is designed for thin but large tissue slices (several cm in lateral size, 0.5mm to 1mm in thickness).
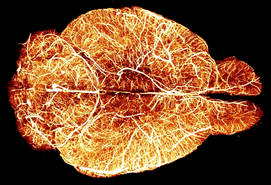
Within the facility hub, LENS can provide the full service (clearing, imaging and processing) or can host external scientists that wish to use the microscopes or acquire technical skills in light-sheet microscopy. Specific projects need to be discussed beforehand to ensure an optimal planning of resource usage and the organisation of specific training if needed.
LENS is an interdisciplinary international laboratory doing research in the field of biophotonics, photonic materials, and quantum technologies. It is home to a PhD school and offers the International Doctorate in Atomic and Molecular Photonics through the University of Florence.
Additional information: bio.lens.unifi.it
Competence Centres
The HUB is based in UNIPV and performs multiscale modelling spanning from neurons and microcircuits to large-scale networks and the whole brain. The HUB leverages on the unique concept of the Brain Scaffold Builder (BSB), a modelling workflow that allows multiscale modeling of neurons, microcircuits, large-scale networks and whole brains.

Although initially designed for the cerebellum, BSB can be easily applied to any brain networks through customised developments on demand. The BSB supports different modelling platforms and formats and can be applied to neuromorphic hardware, robotic control systems and virtual brain simulators. A special aspect of the HUB is to provide the infrastructure and expertise for multiscale experimental validation of the models in rodents. The HUB is relying on EBRAINS and shares with it a set of highly integrated APIs and several sophisticated model building tools.
Dedicated community managers will help users through any issue they may have in exploiting the HUB, in arranging the specific development workflows and simulation of interest, or to help in getting the appropriate HPC allocation for the project. In summary, the HUB will allow researchers and customers with different interests, background, and expertise to develop their own computational models and transform them into different kinds of applications. Dedicated tutorials and community managers will guide users through all the steps needed to reach their specific goals.
The highest impact of this unique installation is expected on engineering and medicine, especially concerning brain-inspired technologies and the investigation and cure of brain diseases.
Additional information: https://dangelo.unipv.it/
The Virtual Brain (TVB) is a full brain network simulation engine, which significantly reduces the gap between computational models and human brain imaging data. The TVB Facility Hub and competence centre at Aix-Marseille University provides field-specific expert support for activities linked to the use of TVB (model building, simulation, analysis, validation and model inversion). Training is provided in TVB workshops and through internships. The TVB Facility Hub serves as the first point of contact for domain-specific questions and refers users to other HBP/EBRAINS groups for further assistance as appropriate.

Potential users: Students, postdocs and researchers in neuroscience, as well as engineers in neurotechnology and clinicians.
Uniqueness: Field-specific expert performance in various domains (high-performance computing, simulation science, brain imaging, neuroscience) is integrated into complex workflows, enabling interoperability of tools within TVB and EBRAINS eco-system. TVB Facility Hub and competence centre provides the user with support in building up an integrated understanding of TVB and adapting the workflows to the user’s own needs.
Impact: To increase the capacity of neuroscientists to use ICT tools such as TVB and EBRAINS to advance science and enable more efficient translation into clinical applications.
Additional information:
Publications:
Sanz-Leon P, Knock SA, Woodman MM, Domide L, Mersmann J, McIntosh AR, Jirsa VK (2013). The Virtual Brain: a simulator of primate brain network dynamics. Frontiers in Neuroinformatics 7:10. doi: 10.3389/fninf.2013.00010
Sanz-Leon P, Knock SA, Spiegler A, Jirsa VK (2015). Mathematical framework for large-scale brain network modelling in The Virtual Brain. Neuroimage 1;111:385-430. https://doi.org/10.1016/j.neuroimage.2015.01.002
Jirsa V., Woodman M.M., Domide L. (2021). The Virtual Brain (TVB): Simulation Environment for Large-Scale Brain Networks. In: Jaeger D., Jung R. (eds) Encyclopedia of Computational Neuroscience. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-7320-6_100682-1
M. Schirner, L. Domide, D. Perdikis, P. Triebkorn, L. Stefanovski, R. Pai, P. Popa, B. Valean, J. Palmer, C. Langford, A. Blickensdörfer, M. van der Vlag, S. Diaz-Pier, A. Peyser, W. Klijn, D. Pleiter, A. Nahm, O. Schmid, M. Woodman, L. Zehl, J. Fousek, S. Petkoski, L. Kusch, M. Hashemi, D. Marinazzo, J.-F. Mangin, A. Flöel, S. Akintoye, B. C. Stahl, M. Cepic, E. Johnson, G. Deco, A. R. McIntosh, C. C. Hilgetag, M. Morgan, B. Schuller, A. Upton, C. McMurtrie, T. Dickscheid, J. G. Bjaalie, K. Amunts, J. Mersmann, V. Jirsa, and P. Ritter (2021). Brain simulation as a cloud service: The Virtual Brain on EBRAINS. Neuroimage 251. https://doi.org/10.1016/j.neuroimage.2022.118973
ANDA is a research data management service targeting neuroscience researchers who are planning to share and publish their research data on the EBRAINS platform, adhering to the FAIR principles of open science. EBRAINS is a research infrastructure providing services for brain and brain-inspired research, including Data and Knowledge services for sharing, finding, and using research data.
Why
Many national and international funders, agencies, and stakeholders have instituted requirements for sharing of research data. An increasing number of researchers are also seeing the need for data sharing using new technologies that will make their research more productive and competitive. EBRAINS offers solutions to meet these expectations while serving the needs of both the data providers and data users. Data sharing requires planning. ANDA assists at an early stage in the planning towards data sharing.
The EBRAINS Data and Knowledge services
The EBRAINS Data and Knowledge services consist of an integrated suite of services, policies, and practices for Findable, Accessible, Interoperable, and Re-usable (FAIR) data, making it easy for the broader neuroscience community to share and publish data and models, and to find and use heterogeneous data, models, and related software relevant for a broad range of research fields in basic and clinical neuroscience. The services also meet the needs of researchers who are aiming to publish data accompanying peer-reviewed journal articles.
ANDA offers advice on how to develop a Data Management Plan for data sharing on EBRAINS
A key instrument for the planning of data sharing is the formalized Data Management Plan. ANDA provides advice on how to develop a Data Management Plan tailored for sharing through EBRAINS. Using ANDA ensures that the management and sharing of your research data comply with the FAIR principles, making data Findable, Accessible, Interoperable and Reusable.
Key aspects
- How to prepare for data sharing: organizing data and capturing metadata
- How to sort concepts and define roles and responsibilities of research staff
- How to use EBRAINS recommended metadata standards
- How to use EBRAINS curation services
- How to publish data and create data collections on EBRAINS
- How to prepare for use of EBRAINS services for data analysis
Who can use our service?
Neuroscientist from around the world and from all fields of neuroscience, ranging from in vitro studies to human brain research, who are aiming to share their data through the EBRAINS Data and Knowledge services.
Contact/ booking information:
You can request ANDA support by filling in this request form https://nettskjema.no/a/207496#/page/1
For general questions about the EBRAINS Data and Knowledge services, please contact support@ebrains.eu
For more information about EBRAINS Data and Knowledge Services, visit https://ebrains.eu/services/data-and-knowledge/
The Virtual Brain Cloud Facility Hub at Charité Berlin offers Brain Simulation as a Service: The Virtual Brain on EBRAINS. Services are being made available as open-source cloud ecosystem on EBRAINS: https://ebrains.eu/service/the-virtual-brain.
Services comprise software for constructing, simulating and analysing brain network models on EBRAINS cloud and FENIX-ICEI infrastructure in compliance with data protection law. The service includes the following components:
- TVB network simulator
- Magnetic resonance imaging processing pipelines to extract structural and functional connectomes
- Multiscale co-simulation of spiking and large-scale networks
- Simulation-ready brain network models of patients and healthy volunteers
- Interfaces to knowledge bases (ScaiView literature mining, pathway inventory NeuroMMSig, EBRAINS atlases) for the integration of biological knowledge in 3D-brain models
- Extensive educational material
- Data protection concept & data protection impact assessment
TVB cloud services facilitate reproducible online collaboration and discovery of data assets, models, and software embedded in scalable and secure workflows, a precondition for research on large cohort data sets, better generalisability and clinical translation.
The Integrated TVB-EBRAINS Services are an outcome of the Co-Design Project The Virtual Brain under lead of Prof. Dr. Petra Ritter (Charité) during SGA2 - and in collaboration with the European Open Science Cloud (EOSC) Project Virtual Brain Cloud under lead of Prof. Dr. Petra Ritter (Charité) - a Partnering Project of the HBP. Services are being further developed and extended during SGA3 as part of Tasks in HBP Work Packages 1, 4, 5, 6 and HBP's Service for Sensitive Data – EBRAINS Health Data Cloud: https://www.healthdatacloud.eu/
Additional information: https://www.brainsimulation.org/, https://ebrains.eu/service/the-virtual-brain
Publications:
SM. Schirner, L. Domide, D. Perdikis, P. Triebkorn, L. Stefanovski, R. Pai, P. Popa, B. Valean, J. Palmer, C. Langford, A. Blickensdörfer, M. van der Vlag, S. Diaz-Pier, A. Peyser, W. Klijn, D. Pleiter, A. Nahm, O. Schmid, M. Woodman, L. Zehl, J. Fousek, S. Petkoski, L. Kusch, M. Hashemi, D. Marinazzo, J.-F. Mangin, A. Flöel, S. Akintoye, B. C. Stahl, M. Cepic, E. Johnson, G. Deco, A. R. McIntosh, C. C. Hilgetag, M. Morgan, B. Schuller, A. Upton, C. McMurtrie, T. Dickscheid, J. G. Bjaalie, K. Amunts, J. Mersmann, V. Jirsa, and P. Ritter (2021). Brain simulation as a cloud service: The Virtual Brain on EBRAINS. Neuroimage 251. https://doi.org/10.1016/j.neuroimage.2022.118973
Schirner, McIntosh, Jirsa, Deco, Ritter (2018). Inferring multi-scale neural mechanisms with brain network modelling. eLife. https://doi.org/10.7554/eLife.28927
Schirner, M., S. Rothmeier, V. Jirsa, A. R. McIntosh and Ritter, P. (2015). An automated pipeline for constructing personalised virtual brains from multimodal neuroimaging data. Neuroimage 117. https://doi.org/10.1016/j.neuroimage.2015.03.055
Ritter, P., M. Schirner, A. R. McIntosh and V. K. Jirsa (2013). The virtual brain integrates computational modeling and multimodal neuroimaging. Brain Connect 3(2): 121-145. https://doi.org/10.1089/brain.2012.0120
Stefanovski, Triebkorn, Spiegler, Diaz-Cortes, Solodkin, Jirsa, McIntosh, Ritter; for the Alzheimer’s Disease Neuroimaging Initiative (2019). Linking molecular pathways and large-scale computational modeling to assess candidate disease mechanisms and pharmacodynamics in Alzheimer’s disease. Frontiers Computational Neuroscience. https://doi.org/10.3389/fncom.2019.00054
Meier, Perdikis, Blickensdörfer, Stefanovski, Liu, Maith, Dinkelbach, Baladron, Hamker, Ritter (2021). Virtual deep brain stimulation: Multiscale co-simulation of spiking basal ganglia model and whole-brain mean-field model with The Virtual Brain Experimental Neurology. https://doi.org/10.1016/j.expneurol.2022.114111 and (BioRxiv) https://doi.org/10.1101/2021.05.05.442704
The Hippocampus Hub is a Reference and Community Web Portal for scientists, students, and interested users to explore data and use, or implement, models related to the hippocampus.
In the Reference Model section, biophysically detailed models of neurons, synapses and a hippocampal microcircuit are featured. The hippocampus CA1 will follow in 2022. This section allows the user to review the models in detail, explore the underlying data and execute models in in silico experiments, using high performance computing infrastructure provided by the HBP EBRAINS infrastructure.
In the Community Model section, an interoperable database of hippocampal related data, such as morphologies or electrophysiological traces are fetched from public resources and a modelling portal for single cell and circuit in silico experiments is provided.
The Hippocampus Hub is a unique facility and we expect to have a big impact on the consolidation and reach of dissemination for hippocampus research, helping to increase the community uptake.
Additional information: Hippocampus Hub website
The Basal Ganglia Modeling Center in Stockholm is co-hosted at the Royal Institute of Technology (KTH) and Karolinska Institute (KI). This modelling Hub facilitates modelling and simulations of the basal ganglia, using a data-driven approach. Modelling is performed at multiple scales, from the molecular, detailed neuron and microcircuit-level, to large scale brain networks and behaviour, where both function and dynamics can be linked between the scales and investigated.
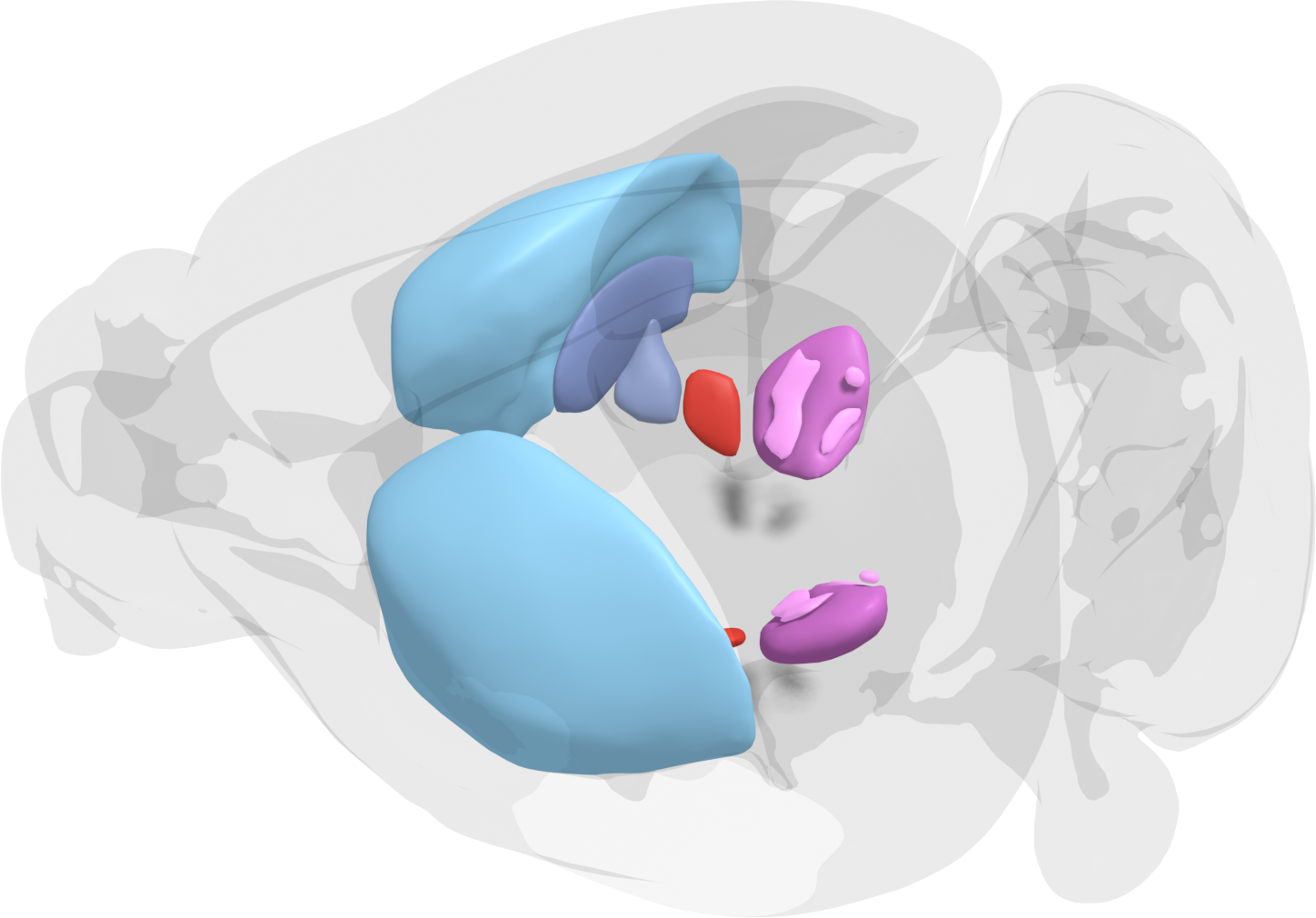
Emphasis is on allowing the user to set up new or use pre-defined virtual experiments to investigate the basal ganglia and related structures. These virtual experiments can then be directly compared to, or verified by, animal experiments. As the models are refined and allow a test of putative mechanisms for learning and selection of behaviour, this will lead to new insights and reduce the number of animals needed for experiments.
The Hub is relying on EBRAINS tools and workflows. For example, a software framework, Snudda, can be used to automatically set up a microcircuit model using detailed morphologically reconstructed neuron models, which is then currently simulated using NEURON on either local machines or on supercomputers for the large networks. Also, a toolset for the simulations of subcellular signalling processes, involved for instance in learning is provided.
Community managers will help users to use and collaborate using the EBRAINS tools and workflows useful for understanding the basal ganglia in health and disease. In addition, some guidance in getting the appropriate HPC allocation for the project will be provided. In summary, the Hub aims at facilitating, for researchers with computational, biological or medical backgrounds and areas of expertise, collaborations around questions regarding basal ganglia function. Tutorials will be provided when needed. Our aim is to create a multidisciplinary community, where scientists help each other, and share their new findings and ideas, forming new collaborations. We will also actively develop open source tools and improve our modelling pipeline.
The highest impact of the Basal Ganglia Modeling Center will likely be on basic science, engineering and medicine, the latter with regards to the many neurological and psychiatric diseases that affect the basal ganglia such as Parkinson’s disease.
Additional info on Basal Ganglia research in Sweden, see SWEBAGS (https://swebags.ebrains.se/) and the International Neuroinformatics Coordinating Facility (https://www.incf.org/)
NEST is a simulator for spiking neural network models based on over 25 years of development that focuses on the dynamics and structure of neural systems, rather than the exact morphology of individual neurons. It is ideal for networks of any size including models of information processing (e.g. in the visual or auditory cortex of mammals), models of network activity dynamics (e.g. laminar cortical networks or balanced random networks) and models of learning and plasticity.
In addition to advanced neuroscientific research, NEST can also be used in education and teaching (NEST Desktop) and as a reference implementation to validate neuromorphic systems.

The NEST Facility Hub, operated by the Forschungszentrum Jülich GmbH, Germany, represents an organizational point of contact that can be approached both from the NEST user and developer side, as well as from the infrastructure side. The Hub offers an up-to-date simulation engine on the level of neurons and synapses provided via Jupyter notebooks on EBRAINS with connection to High Performance Computing (HPC). Additionally it supports users, developers and infrastructure operators in making workflows possible, sustainable and maintainable, acting as a central point bridging between these different views. Users and developers additionally will be provided with software packages for common distributions, as well as containerized solutions such as Docker containers and VM images.
The NEST support helps users of the simulation engine with the installation and handling of the tool and developers in enhancing the openly available community code. For this purposes, a mailing list, an EBRAINS ticketing system, a fortnightly open NEST developers video conference, a Github issue tracker for the coordination of activities, and code reviews have been set up (see https://www.nest-simulator.org/community/). User and developer engagement is supported by user-level documentation, developer documentation and maintenance, and operations documentation (see https://nest-simulator.readthedocs.io/en/v3.3/).
Additional information:
- Homepage: https://www.nest-simulator.org
- Community: https://nest-simulator.org/community/
- EBRAINS: https://ebrains.eu/service/nest-simulator/
- EBRAINS ticketing system: https://ebrains.eu/support/



