Whole Mouse Brain Modelling Workflow
Resources
The first versions of the cerebro-cortical-cerebellar closed loops models will be available on the Brain Simulation Platform from April 2020.
In detail:
- closed-loop modelling and validation of large scale cerebro-cerebellar loops
- olivocerebellar Spiking Neural Network with advanced point neuron models capturing essential non-linear neuronal dynamics
- a tandem model of the cerebellum (forward + inverse cerebellar modules) has been implemented by means of a realistic point-neuron Spiking Neural Network, embedded with distributed plasticity
- model and simulate the forward and inverse controller functions of the cerebro-cortical-cerebellar loops in a closed-loop setting, emulating error-based learning and action prediction
- implementation of a closed-loop experiment (the Vestibulo-Ocular-Reflex) controlled by a cerebellum model in the Neurorobotics Platform
You can access them via the following Collaboratory links:
Showcase - Cerebellum in the NRP
Cerebro-cerebellar closed-loop experiment
Hackathon on Cerebellum Modelling 2020
Publications
Publications
A list of recent and relevant publications:
-
Antonietti A., Martina D., Casellato C., D’Angelo E. and Pedrocchi A., “Control of a Humanoid NAO Robot by an Adaptive Bioinspired Cerebellar Module in 3D Motion Tasks”, Computational Intelligence and Neuroscience, vol. 2019, Article ID 4862157, 15 pages, 2019. https://doi.org/10.1155/2019/4862157
-
Casali S., Marenzi E., Medini C., Casellato C., D’Angelo E., “Reconstruction and Simulation of a Scaffold Model of the Cerebellar Network”, Frontiers in Neuroinformatics Vol 13, pags 37; 19/05/2019; https://doi.org/10.3389/fninf.2019.00037
-
Geminiani A., Pedrocchi A., D’Angelo E., Casellato C., “Response Dynamics in an Olivocerebellar Spiking Neural Network With Non-linear Neuron Properties. Front Comput Neurosci. 2019 Oct 1;13:68. doi: 10.3389/fncom.2019.00068.
__________________________
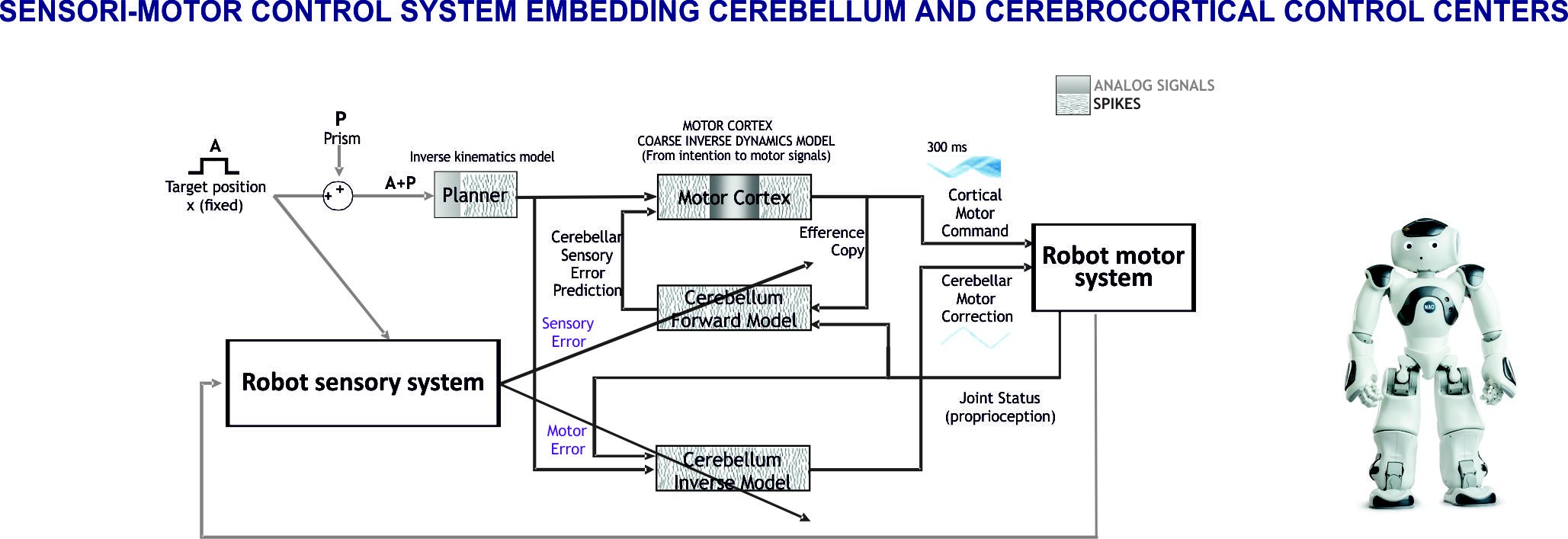
Fig. 1. Sensori-motor control system embedding cerebellum and cerebrocortical control centers
The loops formed by the cerebral cortex and cerebellum passing through the thalamus and other intercalated nuclei are of great importance for their involvement in sensori-motor and cognitive control. These loops are at the core of the predictive ability of the brain and address therefore the intimate mechanism of whole brain functioning. Given the remarkable advancements in cerebral cortical and cerebellar modeling, we have envisaged that models of the cerebrocortical-cerebellar loops are the starting point for building whole brain models. This approach allows not just to face core brain theories, but also to establish the required modeling techniques and tools and to prepare the ground for a multi-modular expansion toward whole-brain models. We have started with a computational architecture employing a tandem configuration of forward and inverse internal models that is known to enable efficient learning in the cerebellum. The cerebellar modules are inserted in a modular system architecture reproducing functional adaptive loops. Furthermore, this is a key element for robotic controllers and when scaling up and mapping the system in an atlas-based whole brain simulator. The cerebellar microcircuit with detailed structure and dynamics becomes thus part of a complex system, fundamental to understand the mutual roles of the different modules and different neural regions.
What makes whole brain modelling special?
The whole brain loops are fundamental to understand the interactive roles among multiple brain regions and modules. A system architecture able to host multiple models and connect them allows to investigate how complex functions emerge. In particular, the cerebrocortical-cerebellum loop is a key element to generate motor, cognitive and emotional control and gives a novel perspective on the mechanisms of implicit learning and memory.
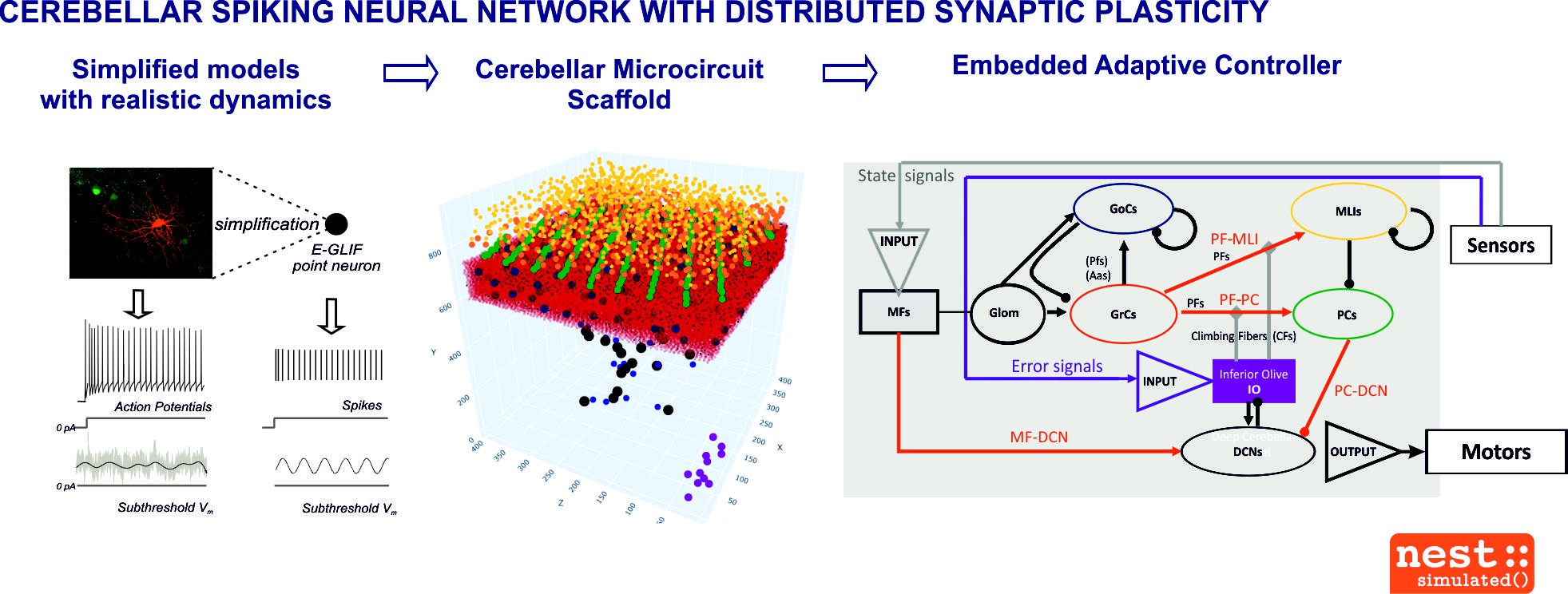
Fig. 2. Cerebellar spiking neural network with distributed synaptic plasticity
What are the specific questions we want to address in the HBP?
The present aim is to model whole brain functionalities by leveraging on the existing microcircuit cerebellar models. We will model, simulate and validate the forward and inverse controller functions of the cerebrocortical-cerebellar loops in a robotic setting, emulating error-based learning and action prediction. The model will be used to simulate specific sensorimotor tasks in closed loop. The models run in pyNEST, are uploaded on the Brain Simulation Platform (BSP) and adapted also for the Neurorobotic Platform (NRP). The work is aligned with future developments and will continue in SGA3 to include more extended and complex circuit representations.
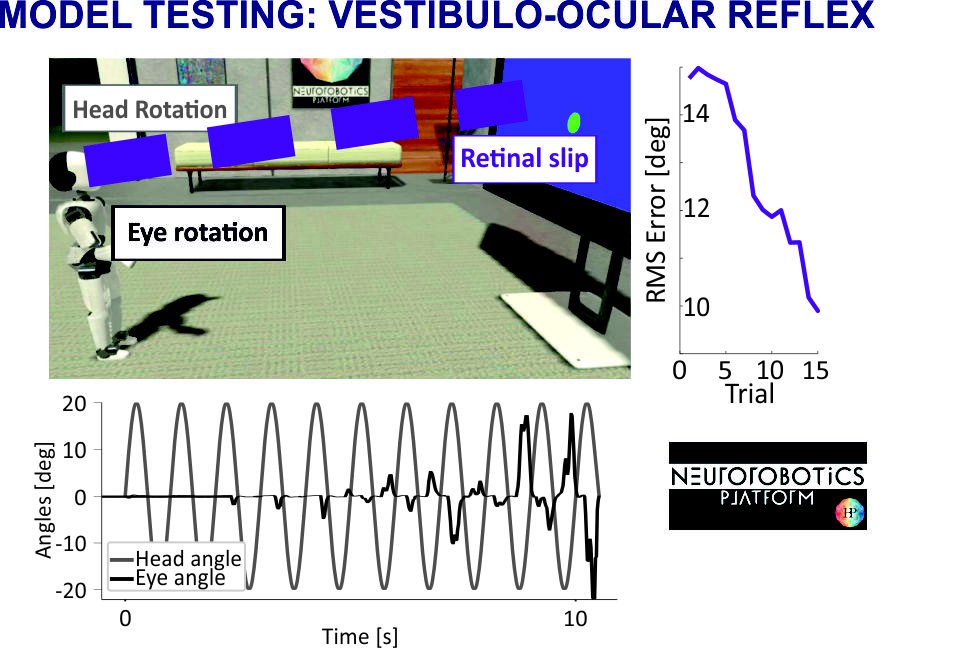
Fig. 3. Model testing: vestibulo-ocular reflex
What is our specific take?
The modular cerebrocortico-cerebellar circuit has been seen as an interesting use-case for different platforms: the BSP (where the cerebrocortico-cerebellar model can be directly launched by the users, eventually exploiting parallel computing) and the NRP (where the same cerebrocortico-cerebellar model can be used as an initial brain model for a neurorobotic plant actuated in a task-dependent environment). The models can be used not only by other HBP partners but also by scientists external to the HBP consortium through simple access pipelines provided by the BSP and NRP. The integration of more biologically inspired components (realistic modelling of cerebral cortex, brainstem, spinal cord and neuromuscular system) is of great interest for several research groups as manifested at the recent Hackathon on Cerebellum Modelling .
Roadmap
In March 2020, as part of the BSP, we have released the first model of a cerebrocortical-cerebellar closed-loop experiment. The aim was to embed plasticity components (such as synaptic learning mechanisms) into the scaffold model and into large-scale brain loops. These BSP models can be used to investigate multiscale cerebellar circuit functions and for applications to neurorobotics. Moreover, these BSP models serve to aggregate the cerebellum modeling community around the BSP and HBP and future EBRAINS.
Who's involved?
Egidio D’Angelo, University of Pavia
Claudia Casellato, University of Pavia
Alberto Antonietti, University of Pavia
Alice Geminiani, University of Pavia
Stefano Casali, University of Pavia
Dimitri Rodarie, Blue Brain Project, École Polytechnique Fédérale de Lausanne
Edoardo Negri, University of Pavia
Robin De Schepper, University of Pavia
Simona Tritto, University of Pavia
Alessandra Pedrocchi and Alessandra Trapani (Politecnico di Milano) are contributing from cerebNEST.
What's in it for me?
Collabs, links, and live papers listed in the resources box above.
Participate in community modelling: we would be happy to hear from you if you would like to get involved and contribute to our community effort. For all information, please contact Simona Tritto by e-mail at simona.tritto@unipv.it



